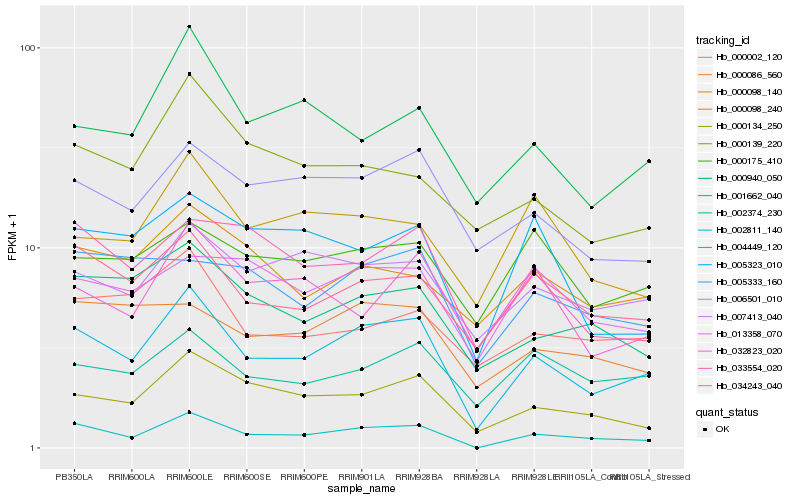
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004449_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631291 [Jatropha curcas] |
| 2 |
Hb_002811_140 |
0.0921646301 |
- |
- |
PREDICTED: uncharacterized protein LOC105647665 [Jatropha curcas] |
| 3 |
Hb_000134_250 |
0.1222028929 |
- |
- |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_034243_040 |
0.1289537672 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor VRN1-like [Jatropha curcas] |
| 5 |
Hb_006501_010 |
0.1333776534 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3A isoform X1 [Populus euphratica] |
| 6 |
Hb_007413_040 |
0.1360585867 |
- |
- |
myo inositol monophosphatase, putative [Ricinus communis] |
| 7 |
Hb_001662_040 |
0.1429274775 |
- |
- |
DNA helicase, putative [Ricinus communis] |
| 8 |
Hb_000940_050 |
0.1440172566 |
- |
- |
hypothetical protein JCGZ_19588 [Jatropha curcas] |
| 9 |
Hb_002374_230 |
0.1445073678 |
- |
- |
PREDICTED: centromere-associated protein E [Jatropha curcas] |
| 10 |
Hb_033554_020 |
0.1476451661 |
transcription factor |
TF Family: BES1 |
PREDICTED: beta-amylase 8 [Jatropha curcas] |
| 11 |
Hb_000098_240 |
0.1502532877 |
- |
- |
PREDICTED: UDP-glucose:glycoprotein glucosyltransferase [Jatropha curcas] |
| 12 |
Hb_000086_560 |
0.1513469983 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR2 isoform X2 [Jatropha curcas] |
| 13 |
Hb_032823_020 |
0.1519004269 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000002_120 |
0.1524785416 |
- |
- |
PREDICTED: uncharacterized protein LOC105782064 [Gossypium raimondii] |
| 15 |
Hb_000098_140 |
0.1526343554 |
- |
- |
hypothetical protein JCGZ_04989 [Jatropha curcas] |
| 16 |
Hb_005323_010 |
0.1531653767 |
- |
- |
PREDICTED: uncharacterized protein LOC105642810 [Jatropha curcas] |
| 17 |
Hb_000175_410 |
0.1535107942 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 56-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_013358_070 |
0.1541558931 |
- |
- |
protein with unknown function [Ricinus communis] |
| 19 |
Hb_000139_220 |
0.1541971464 |
- |
- |
leucine rich repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_005333_160 |
0.1551588039 |
- |
- |
hypothetical protein B456_009G230200 [Gossypium raimondii] |