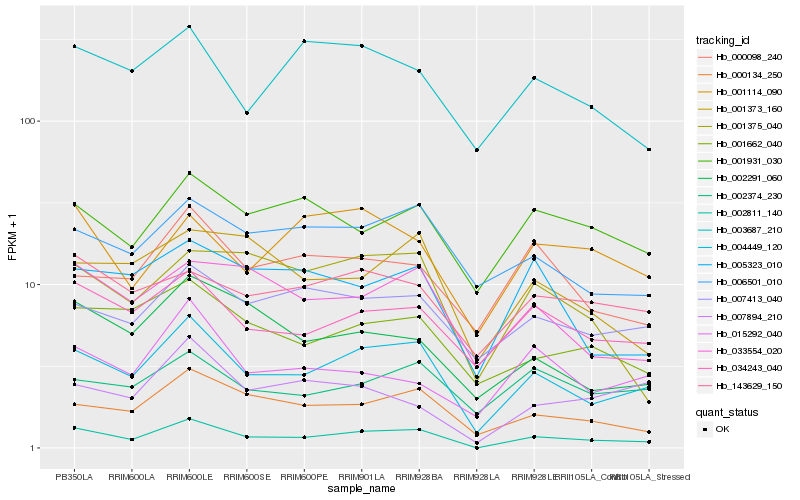
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002811_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647665 [Jatropha curcas] |
| 2 |
Hb_004449_120 |
0.0921646301 |
- |
- |
PREDICTED: uncharacterized protein LOC105631291 [Jatropha curcas] |
| 3 |
Hb_000134_250 |
0.1536677621 |
- |
- |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_034243_040 |
0.1652689927 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor VRN1-like [Jatropha curcas] |
| 5 |
Hb_001662_040 |
0.1753316181 |
- |
- |
DNA helicase, putative [Ricinus communis] |
| 6 |
Hb_033554_020 |
0.1771547567 |
transcription factor |
TF Family: BES1 |
PREDICTED: beta-amylase 8 [Jatropha curcas] |
| 7 |
Hb_002291_060 |
0.1817443028 |
- |
- |
PREDICTED: splicing factor 3A subunit 3-like isoform X1 [Fragaria vesca subsp. vesca] |
| 8 |
Hb_001931_030 |
0.1825418491 |
- |
- |
PREDICTED: T-complex protein 1 subunit eta-like [Nelumbo nucifera] |
| 9 |
Hb_007413_040 |
0.1834975399 |
- |
- |
myo inositol monophosphatase, putative [Ricinus communis] |
| 10 |
Hb_001375_040 |
0.1835242272 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 11 |
Hb_006501_010 |
0.184869181 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3A isoform X1 [Populus euphratica] |
| 12 |
Hb_001114_090 |
0.1851789212 |
- |
- |
PREDICTED: magnesium-dependent phosphatase 1 [Jatropha curcas] |
| 13 |
Hb_005323_010 |
0.1859014912 |
- |
- |
PREDICTED: uncharacterized protein LOC105642810 [Jatropha curcas] |
| 14 |
Hb_003687_210 |
0.1860238218 |
- |
- |
PREDICTED: malate dehydrogenase [Populus euphratica] |
| 15 |
Hb_001373_160 |
0.1883547873 |
- |
- |
hypothetical protein JCGZ_16907 [Jatropha curcas] |
| 16 |
Hb_143629_150 |
0.1885850614 |
- |
- |
PREDICTED: flap endonuclease 1 isoform X2 [Prunus mume] |
| 17 |
Hb_015292_040 |
0.1887255899 |
- |
- |
PREDICTED: myb-like protein X [Jatropha curcas] |
| 18 |
Hb_000098_240 |
0.189578634 |
- |
- |
PREDICTED: UDP-glucose:glycoprotein glucosyltransferase [Jatropha curcas] |
| 19 |
Hb_007894_210 |
0.1903566368 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 22 homolog 1 [Jatropha curcas] |
| 20 |
Hb_002374_230 |
0.1915938057 |
- |
- |
PREDICTED: centromere-associated protein E [Jatropha curcas] |