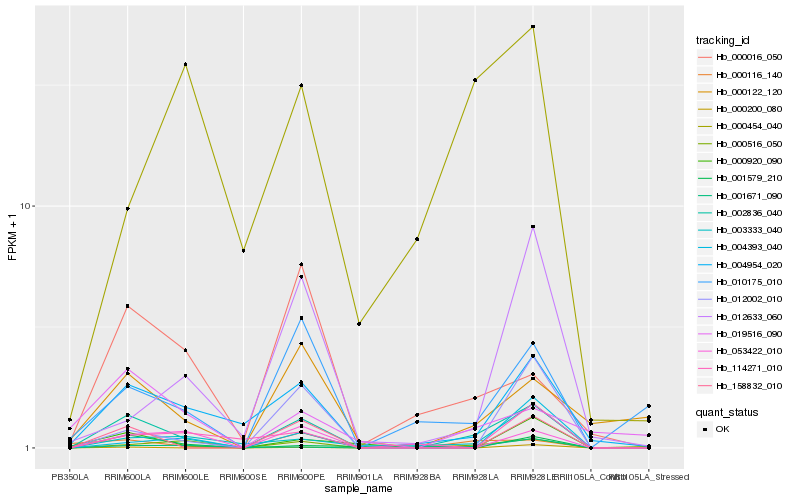
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002836_040 |
0.0 |
- |
- |
Auxin-induced protein 5NG4 [Glycine soja] |
| 2 |
Hb_004954_020 |
0.2683861617 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 51 isoform X2 [Jatropha curcas] |
| 3 |
Hb_158832_010 |
0.2956925558 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000116_140 |
0.3092731366 |
- |
- |
dihydrodipicolinate synthase, putative [Ricinus communis] |
| 5 |
Hb_000516_050 |
0.3128682999 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 6 |
Hb_053422_010 |
0.3181922105 |
- |
- |
PREDICTED: uncharacterized protein LOC105641005 [Jatropha curcas] |
| 7 |
Hb_000122_120 |
0.3323660121 |
- |
- |
PREDICTED: arogenate dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_000920_090 |
0.3332006103 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: vinorine synthase-like [Jatropha curcas] |
| 9 |
Hb_010175_010 |
0.3335646201 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_012002_010 |
0.3378195103 |
- |
- |
hypothetical protein VITISV_016116 [Vitis vinifera] |
| 11 |
Hb_003333_040 |
0.3378322651 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_001579_210 |
0.3392201087 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000016_050 |
0.3543034995 |
- |
- |
PREDICTED: uncharacterized protein LOC105648535 [Jatropha curcas] |
| 14 |
Hb_000200_080 |
0.3611177325 |
- |
- |
PREDICTED: uncharacterized protein LOC104877459 [Vitis vinifera] |
| 15 |
Hb_001671_090 |
0.3617669455 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 16 |
Hb_004393_040 |
0.3698324506 |
transcription factor |
TF Family: RAV |
PREDICTED: AP2/ERF and B3 domain-containing transcription factor At1g50680-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_012633_060 |
0.3743796391 |
- |
- |
laccase, putative [Ricinus communis] |
| 18 |
Hb_000454_040 |
0.3778736963 |
- |
- |
PREDICTED: kinesin-like protein NACK1 [Jatropha curcas] |
| 19 |
Hb_019516_090 |
0.3802683087 |
- |
- |
hypothetical protein POPTR_0004s06350g [Populus trichocarpa] |
| 20 |
Hb_114271_010 |
0.3804451191 |
- |
- |
hypothetical protein JCGZ_19461 [Jatropha curcas] |