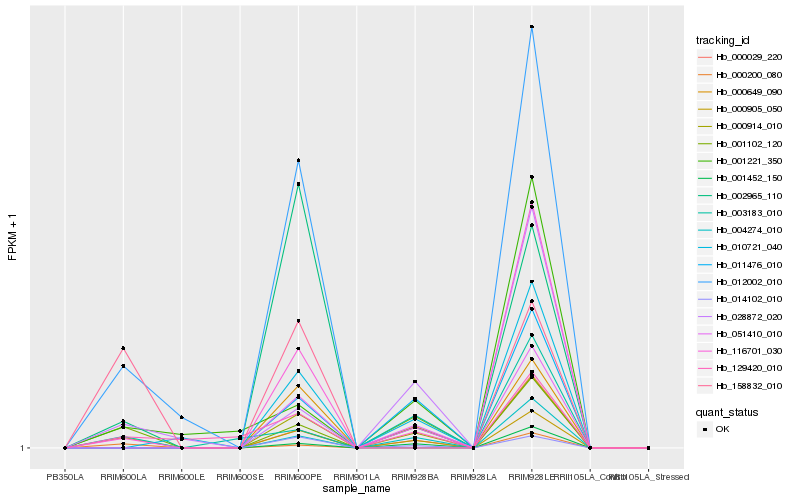
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000905_050 |
0.0 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 2 |
Hb_001102_120 |
0.1442410024 |
- |
- |
- |
| 3 |
Hb_116701_030 |
0.2306785355 |
- |
- |
Pentatricopeptide repeat (PPR-like) superfamily protein [Theobroma cacao] |
| 4 |
Hb_028872_020 |
0.24973948 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 5 |
Hb_129420_010 |
0.2633719303 |
transcription factor |
TF Family: NAC |
NAC transcription factor 059 [Jatropha curcas] |
| 6 |
Hb_003183_010 |
0.2679408428 |
- |
- |
PREDICTED: uncharacterized protein LOC105785048, partial [Gossypium raimondii] |
| 7 |
Hb_000914_010 |
0.2719458111 |
- |
- |
hypothetical protein POPTR_0025s00760g [Populus trichocarpa] |
| 8 |
Hb_051410_010 |
0.2720184703 |
- |
- |
PREDICTED: uncharacterized protein LOC103401813 [Malus domestica] |
| 9 |
Hb_000200_080 |
0.2726564333 |
- |
- |
PREDICTED: uncharacterized protein LOC104877459 [Vitis vinifera] |
| 10 |
Hb_158832_010 |
0.2740717546 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002965_110 |
0.2759557097 |
- |
- |
hypothetical protein POPTR_0010s15040g [Populus trichocarpa] |
| 12 |
Hb_012002_010 |
0.2778789361 |
- |
- |
hypothetical protein VITISV_016116 [Vitis vinifera] |
| 13 |
Hb_000649_090 |
0.2805872255 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_011476_010 |
0.2914235002 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 15 |
Hb_004274_010 |
0.2957023197 |
- |
- |
PREDICTED: uncharacterized protein LOC104451199 [Eucalyptus grandis] |
| 16 |
Hb_001221_350 |
0.2967954754 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_010721_040 |
0.2992916318 |
transcription factor |
TF Family: M-type |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_000029_220 |
0.3016576896 |
- |
- |
PREDICTED: pre-mRNA-splicing factor SLU7-A-like [Jatropha curcas] |
| 19 |
Hb_001452_150 |
0.302245277 |
- |
- |
PREDICTED: uncharacterized protein LOC104879858 [Vitis vinifera] |
| 20 |
Hb_014102_010 |
0.3022543048 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |