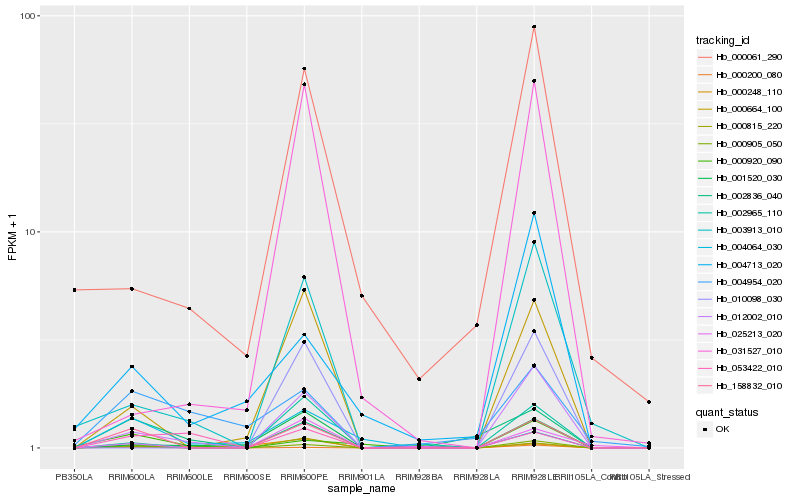
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_158832_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_012002_010 |
0.2493554311 |
- |
- |
hypothetical protein VITISV_016116 [Vitis vinifera] |
| 3 |
Hb_000664_100 |
0.2520243338 |
- |
- |
PREDICTED: uncharacterized protein LOC105638386 [Jatropha curcas] |
| 4 |
Hb_000200_080 |
0.2620808858 |
- |
- |
PREDICTED: uncharacterized protein LOC104877459 [Vitis vinifera] |
| 5 |
Hb_000905_050 |
0.2740717546 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 6 |
Hb_002836_040 |
0.2956925558 |
- |
- |
Auxin-induced protein 5NG4 [Glycine soja] |
| 7 |
Hb_025213_020 |
0.3027176067 |
- |
- |
PREDICTED: (R)-mandelonitrile lyase-like [Populus euphratica] |
| 8 |
Hb_002965_110 |
0.3044864835 |
- |
- |
hypothetical protein POPTR_0010s15040g [Populus trichocarpa] |
| 9 |
Hb_000815_220 |
0.313119965 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000920_090 |
0.3162924311 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: vinorine synthase-like [Jatropha curcas] |
| 11 |
Hb_003913_010 |
0.3208313084 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000248_110 |
0.3462134763 |
- |
- |
hypothetical protein VITISV_037580 [Vitis vinifera] |
| 13 |
Hb_004064_030 |
0.3512307747 |
- |
- |
aldose-1-epimerase, putative [Ricinus communis] |
| 14 |
Hb_001520_030 |
0.3525075866 |
transcription factor |
TF Family: MYB |
PREDICTED: trichome differentiation protein GL1-like isoform X2 [Populus euphratica] |
| 15 |
Hb_053422_010 |
0.3670028012 |
- |
- |
PREDICTED: uncharacterized protein LOC105641005 [Jatropha curcas] |
| 16 |
Hb_004954_020 |
0.368528886 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 51 isoform X2 [Jatropha curcas] |
| 17 |
Hb_031527_010 |
0.3722094295 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000061_290 |
0.3783553283 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004713_020 |
0.378595523 |
- |
- |
- |
| 20 |
Hb_010098_030 |
0.3785962457 |
- |
- |
conserved hypothetical protein [Ricinus communis] |