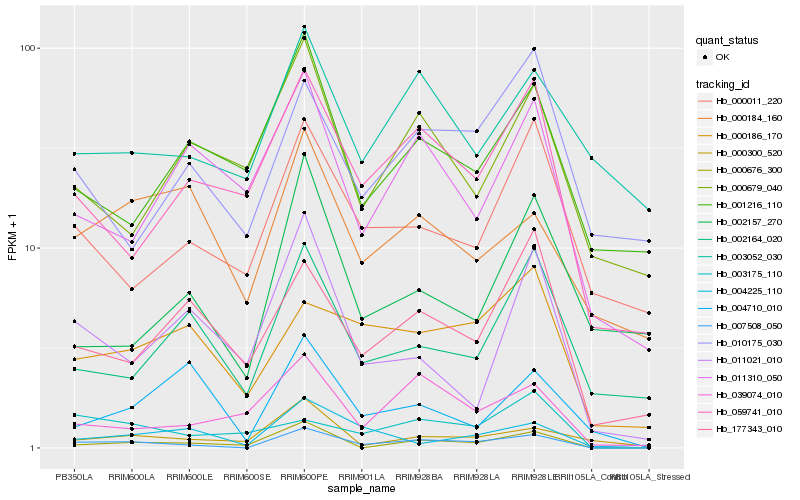
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007508_050 |
0.0 |
- |
- |
PREDICTED: phosphoserine phosphatase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000676_300 |
0.2340472655 |
- |
- |
PREDICTED: probable caffeine synthase 4 [Jatropha curcas] |
| 3 |
Hb_059741_010 |
0.2468950315 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 1 [UDP-forming] [Jatropha curcas] |
| 4 |
Hb_039074_010 |
0.254020216 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 5 |
Hb_000186_170 |
0.265779713 |
- |
- |
pyrophosphate-dependent phosphofructokinase, partial [Hevea brasiliensis] |
| 6 |
Hb_177343_010 |
0.2675955805 |
- |
- |
PREDICTED: uncharacterized protein LOC105650963 [Jatropha curcas] |
| 7 |
Hb_000011_220 |
0.2689253559 |
- |
- |
PREDICTED: uncharacterized protein LOC105631313 [Jatropha curcas] |
| 8 |
Hb_000300_520 |
0.2716016665 |
- |
- |
hypothetical protein JCGZ_02702 [Jatropha curcas] |
| 9 |
Hb_003175_110 |
0.2735816894 |
- |
- |
- |
| 10 |
Hb_002164_020 |
0.275474272 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_011310_050 |
0.2755860847 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 1 [UDP-forming] [Jatropha curcas] |
| 12 |
Hb_004225_110 |
0.2773090374 |
- |
- |
PREDICTED: ABC transporter G family member 28 [Jatropha curcas] |
| 13 |
Hb_011021_010 |
0.2788875661 |
- |
- |
kinase, putative [Ricinus communis] |
| 14 |
Hb_000184_160 |
0.2789055123 |
- |
- |
PREDICTED: uncharacterized protein LOC105647991 [Jatropha curcas] |
| 15 |
Hb_003052_030 |
0.2794938142 |
- |
- |
6-phosphogluconate dehydrogenase family protein isoform 1 [Theobroma cacao] |
| 16 |
Hb_010175_030 |
0.2798189802 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Jatropha curcas] |
| 17 |
Hb_001216_110 |
0.2819253938 |
- |
- |
PREDICTED: protein GPR107-like [Jatropha curcas] |
| 18 |
Hb_000679_040 |
0.2839610608 |
- |
- |
Cellulose synthase 1 [Theobroma cacao] |
| 19 |
Hb_002157_270 |
0.2885198046 |
- |
- |
PREDICTED: transmembrane protein 87B [Jatropha curcas] |
| 20 |
Hb_004710_010 |
0.2887033692 |
- |
- |
hypothetical protein EUGRSUZ_D010351, partial [Eucalyptus grandis] |