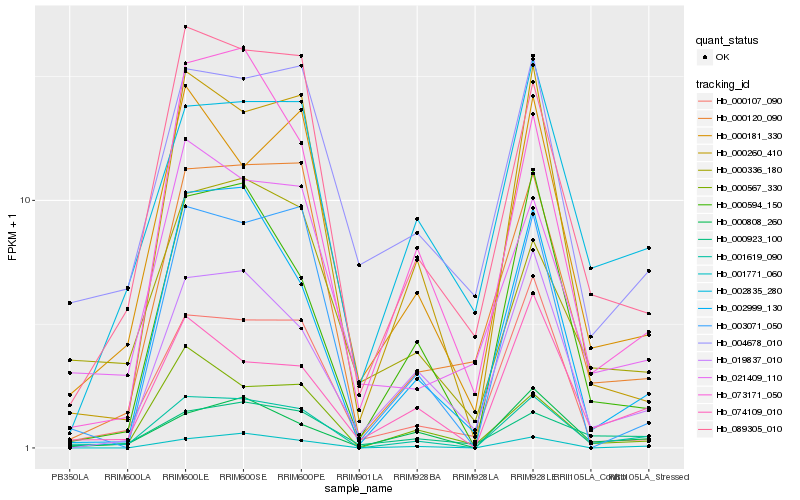
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001619_090 |
0.0 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 2 |
Hb_089305_010 |
0.1566897155 |
- |
- |
PREDICTED: U-box domain-containing protein 26-like [Jatropha curcas] |
| 3 |
Hb_000181_330 |
0.1608305849 |
- |
- |
ubiquitin ligase protein cop1, putative [Ricinus communis] |
| 4 |
Hb_074109_010 |
0.1617337099 |
- |
- |
PREDICTED: putative auxin efflux carrier component 5 [Jatropha curcas] |
| 5 |
Hb_000120_090 |
0.1644307979 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 4-like [Jatropha curcas] |
| 6 |
Hb_002999_130 |
0.1654037826 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0008s08740g [Populus trichocarpa] |
| 7 |
Hb_000808_260 |
0.1672083232 |
- |
- |
hypothetical protein RCOM_1434920 [Ricinus communis] |
| 8 |
Hb_000260_410 |
0.1678265222 |
- |
- |
PREDICTED: protein LURP-one-related 8 [Jatropha curcas] |
| 9 |
Hb_000594_150 |
0.1679319689 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN7, chloroplastic-like isoform X1 [Nelumbo nucifera] |
| 10 |
Hb_001771_060 |
0.1740894814 |
- |
- |
hypothetical protein B456_012G144100 [Gossypium raimondii] |
| 11 |
Hb_000107_090 |
0.1746859093 |
- |
- |
PREDICTED: kinesin KP1 [Jatropha curcas] |
| 12 |
Hb_000923_100 |
0.1775402741 |
- |
- |
chloride channel clc, putative [Ricinus communis] |
| 13 |
Hb_002835_280 |
0.1775802126 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.1 [Jatropha curcas] |
| 14 |
Hb_021409_110 |
0.1779997162 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 15 |
Hb_019837_010 |
0.178025395 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004678_010 |
0.1785170616 |
transcription factor |
TF Family: Orphans |
Salt-tolerance protein, putative [Ricinus communis] |
| 17 |
Hb_003071_050 |
0.1820677515 |
transcription factor |
TF Family: ERF |
ethylene-responsive element binding protein 2 [Hevea brasiliensis] |
| 18 |
Hb_000567_330 |
0.1823799306 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL7 [Jatropha curcas] |
| 19 |
Hb_073171_050 |
0.1825402376 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein H [Jatropha curcas] |
| 20 |
Hb_000336_180 |
0.1831934207 |
- |
- |
conserved hypothetical protein [Ricinus communis] |