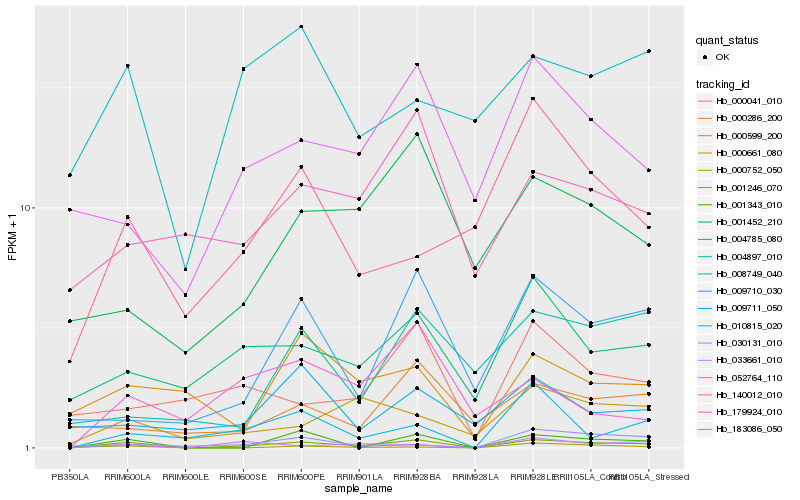
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001246_070 |
0.0 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001343_010 |
0.2450601114 |
- |
- |
- |
| 3 |
Hb_010815_020 |
0.283906588 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_009710_030 |
0.2851599613 |
- |
- |
- |
| 5 |
Hb_004785_080 |
0.2875866796 |
- |
- |
hypothetical protein POPTR_0005s21250g [Populus trichocarpa] |
| 6 |
Hb_030131_010 |
0.2920730691 |
- |
- |
PREDICTED: probable acyl-activating enzyme 1, peroxisomal, partial [Jatropha curcas] |
| 7 |
Hb_052764_110 |
0.2976440925 |
- |
- |
PREDICTED: quinone oxidoreductase PIG3 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000661_080 |
0.2988701964 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 9 |
Hb_000752_050 |
0.3008298754 |
- |
- |
PREDICTED: uncharacterized protein LOC102610887 [Citrus sinensis] |
| 10 |
Hb_000286_200 |
0.3030958719 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_000041_010 |
0.3038812542 |
- |
- |
pfkB-type carbohydrate kinase family protein [Populus trichocarpa] |
| 12 |
Hb_000599_200 |
0.305012409 |
- |
- |
ABC transporter family protein [Populus trichocarpa] |
| 13 |
Hb_001452_210 |
0.3055904664 |
- |
- |
molybdopterin biosynthesis CNX1 protein [Arabidopsis thaliana] |
| 14 |
Hb_009711_050 |
0.3100884608 |
- |
- |
hypothetical protein POPTR_0019s04500g [Populus trichocarpa] |
| 15 |
Hb_008749_040 |
0.3128381003 |
- |
- |
PREDICTED: formin-like protein 20 [Populus euphratica] |
| 16 |
Hb_179924_010 |
0.3128751822 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62930, chloroplastic-like [Populus euphratica] |
| 17 |
Hb_140012_010 |
0.3163098127 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Eucalyptus grandis] |
| 18 |
Hb_183086_050 |
0.3177184875 |
- |
- |
PREDICTED: metal-nicotianamine transporter YSL3 [Jatropha curcas] |
| 19 |
Hb_033661_010 |
0.3188005585 |
- |
- |
PREDICTED: uncharacterized protein LOC105852393, partial [Cicer arietinum] |
| 20 |
Hb_004897_010 |
0.3192326891 |
- |
- |
- |