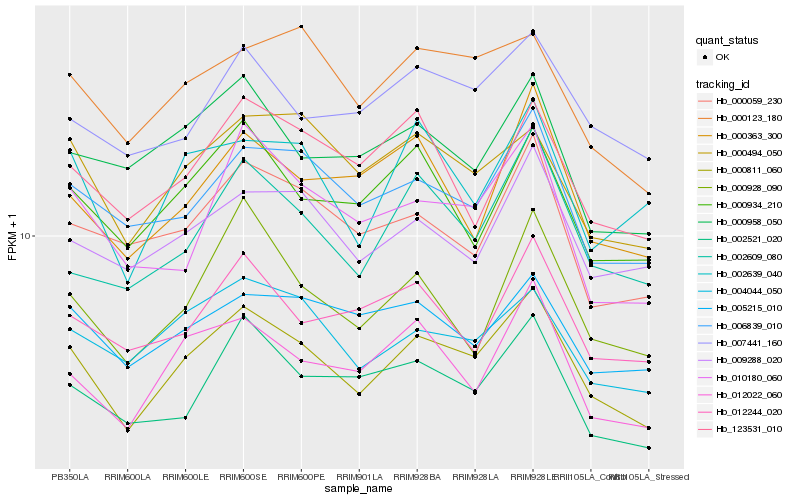
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000811_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650696 [Jatropha curcas] |
| 2 |
Hb_012022_060 |
0.0850059665 |
- |
- |
PREDICTED: uncharacterized protein LOC105646588 isoform X2 [Jatropha curcas] |
| 3 |
Hb_006839_010 |
0.0861077345 |
- |
- |
PREDICTED: importin subunit beta-1-like [Jatropha curcas] |
| 4 |
Hb_004044_050 |
0.0873497355 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Jatropha curcas] |
| 5 |
Hb_000494_050 |
0.090498925 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 6 |
Hb_002609_080 |
0.0907807362 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 7, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_000123_180 |
0.0914614056 |
- |
- |
2-oxoglutarate dehydrogenase, putative [Ricinus communis] |
| 8 |
Hb_000934_210 |
0.0924342908 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 9 |
Hb_002521_020 |
0.0935454967 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g03100, mitochondrial-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_002639_040 |
0.0936587517 |
- |
- |
PREDICTED: T-complex protein 1 subunit beta [Jatropha curcas] |
| 11 |
Hb_009288_020 |
0.0940648315 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 12 |
Hb_010180_060 |
0.0941831509 |
- |
- |
PREDICTED: uncharacterized protein LOC105648049 [Jatropha curcas] |
| 13 |
Hb_007441_160 |
0.0952492942 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_000059_230 |
0.0956142291 |
- |
- |
PREDICTED: uncharacterized protein LOC105648049 [Jatropha curcas] |
| 15 |
Hb_000928_090 |
0.0969836964 |
- |
- |
PREDICTED: septin and tuftelin-interacting protein 1 homolog 1 [Jatropha curcas] |
| 16 |
Hb_000958_050 |
0.0987609647 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |
| 17 |
Hb_000363_300 |
0.0989339515 |
- |
- |
PREDICTED: protein ENHANCED DOWNY MILDEW 2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_005215_010 |
0.0993642883 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_123531_010 |
0.0995666543 |
- |
- |
PREDICTED: nuclear pore complex protein NUP98A isoform X1 [Jatropha curcas] |
| 20 |
Hb_012244_020 |
0.1001692388 |
- |
- |
calpain, putative [Ricinus communis] |