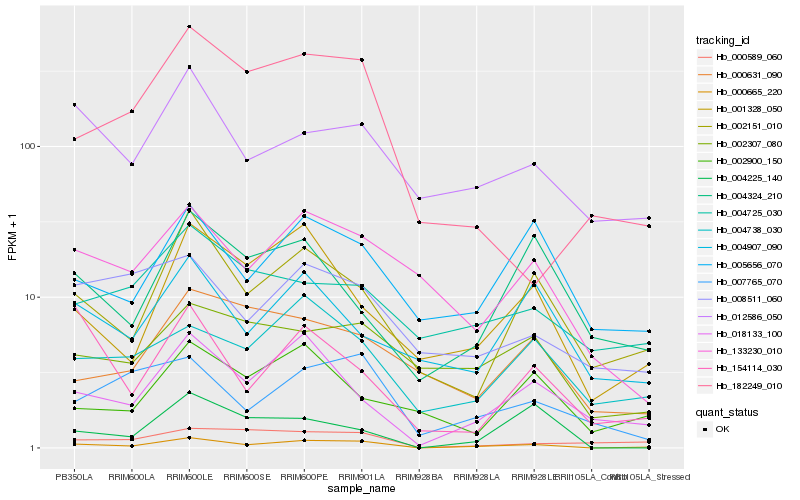
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000665_220 |
0.0 |
- |
- |
catalytic, putative [Ricinus communis] |
| 2 |
Hb_002151_010 |
0.2035550278 |
- |
- |
hypothetical protein POPTR_0006s14610g [Populus trichocarpa] |
| 3 |
Hb_018133_100 |
0.207652728 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_001328_050 |
0.2216434302 |
- |
- |
PREDICTED: uncharacterized protein LOC105640854 [Jatropha curcas] |
| 5 |
Hb_005656_070 |
0.2231927125 |
- |
- |
PREDICTED: cryptochrome DASH, chloroplastic/mitochondrial [Jatropha curcas] |
| 6 |
Hb_004738_030 |
0.2256744415 |
- |
- |
Inactive protein kinase [Gossypium arboreum] |
| 7 |
Hb_133230_010 |
0.2316632267 |
- |
- |
hypothetical protein L484_001846 [Morus notabilis] |
| 8 |
Hb_012586_050 |
0.2332935163 |
- |
- |
PREDICTED: triosephosphate isomerase, cytosolic [Prunus mume] |
| 9 |
Hb_007765_070 |
0.2336527243 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 2 [Jatropha curcas] |
| 10 |
Hb_002307_080 |
0.24200966 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004225_140 |
0.2479589215 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g33260 [Jatropha curcas] |
| 12 |
Hb_004907_090 |
0.2495663074 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_008511_060 |
0.2516931072 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At4g34500 [Jatropha curcas] |
| 14 |
Hb_000589_060 |
0.2537533572 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_00699 [Jatropha curcas] |
| 15 |
Hb_004324_210 |
0.2556529459 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 16 |
Hb_154114_030 |
0.2559171915 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 17 |
Hb_182249_010 |
0.2574276969 |
- |
- |
- |
| 18 |
Hb_002900_150 |
0.258253314 |
- |
- |
PREDICTED: syntaxin-81 [Jatropha curcas] |
| 19 |
Hb_000631_090 |
0.2591024968 |
- |
- |
PREDICTED: uncharacterized protein LOC105640619 [Jatropha curcas] |
| 20 |
Hb_004725_030 |
0.2594264266 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X4 [Jatropha curcas] |