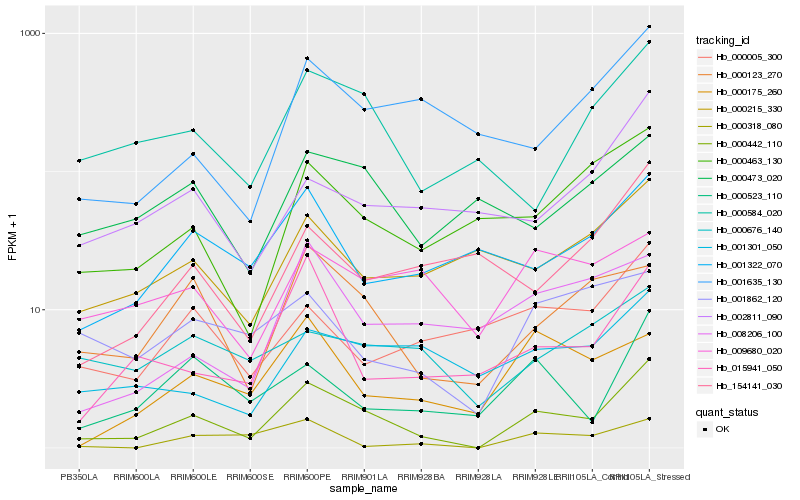
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000442_110 |
0.0 |
- |
- |
hypothetical protein POPTR_0010s18960g [Populus trichocarpa] |
| 2 |
Hb_000123_270 |
0.239078785 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000584_020 |
0.2444003397 |
- |
- |
hypothetical protein PHAVU_002G233500g, partial [Phaseolus vulgaris] |
| 4 |
Hb_015941_050 |
0.2463381975 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000523_110 |
0.2464000222 |
- |
- |
PREDICTED: probable polyamine oxidase 4 [Jatropha curcas] |
| 6 |
Hb_000463_130 |
0.2482708927 |
- |
- |
unnamed protein product [Coffea canephora] |
| 7 |
Hb_001635_130 |
0.2529964724 |
- |
- |
PREDICTED: 60S ribosomal protein L28-2-like [Jatropha curcas] |
| 8 |
Hb_002811_090 |
0.2592175708 |
- |
- |
40S ribosomal protein S24A [Hevea brasiliensis] |
| 9 |
Hb_001301_050 |
0.262750167 |
- |
- |
- |
| 10 |
Hb_001322_070 |
0.262758869 |
- |
- |
protein with unknown function [Ricinus communis] |
| 11 |
Hb_000005_300 |
0.2673726688 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000215_330 |
0.2674632647 |
- |
- |
PREDICTED: AP-4 complex subunit sigma [Jatropha curcas] |
| 13 |
Hb_000175_260 |
0.270268972 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 14-like [Populus euphratica] |
| 14 |
Hb_154141_030 |
0.2712191031 |
- |
- |
PREDICTED: histone H2AX [Populus euphratica] |
| 15 |
Hb_000318_080 |
0.271542169 |
- |
- |
PREDICTED: la-related protein 6C [Jatropha curcas] |
| 16 |
Hb_008206_100 |
0.2733135855 |
- |
- |
PREDICTED: uncharacterized protein LOC105638302 [Jatropha curcas] |
| 17 |
Hb_000676_140 |
0.2734076067 |
- |
- |
structural constituent of ribosome, putative [Ricinus communis] |
| 18 |
Hb_001862_120 |
0.2735495237 |
- |
- |
PREDICTED: aspartic proteinase-like protein 2 [Jatropha curcas] |
| 19 |
Hb_000473_020 |
0.2753820717 |
- |
- |
expressed protein, putative [Ricinus communis] |
| 20 |
Hb_009680_020 |
0.2763124202 |
- |
- |
PREDICTED: 40S ribosomal protein S10 [Jatropha curcas] |