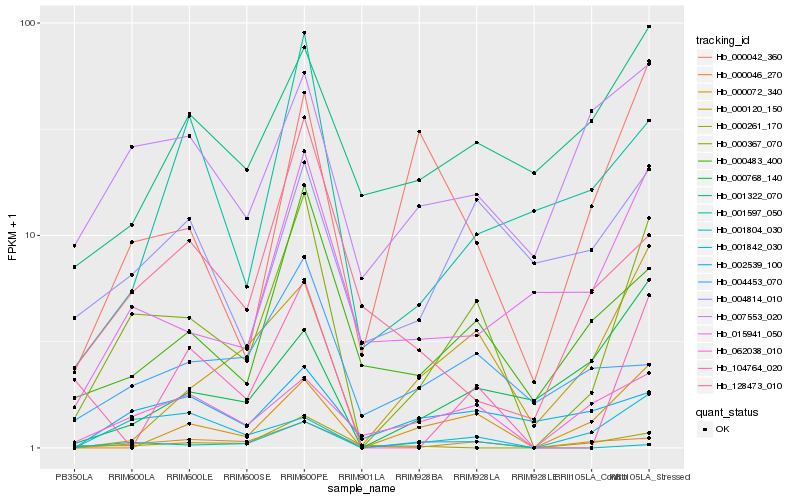
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000367_070 |
0.0 |
- |
- |
hypothetical protein JCGZ_11697 [Jatropha curcas] |
| 2 |
Hb_000046_270 |
0.2551974466 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 2-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_000483_400 |
0.2590245829 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 1, mitochondrial [Jatropha curcas] |
| 4 |
Hb_062038_010 |
0.2644479063 |
transcription factor |
TF Family: B3 |
PREDICTED: uncharacterized protein LOC104882266 isoform X2 [Vitis vinifera] |
| 5 |
Hb_000072_340 |
0.2767980373 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_015941_050 |
0.2781075 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001842_030 |
0.2796304458 |
- |
- |
PREDICTED: histone deacetylase 9 [Jatropha curcas] |
| 8 |
Hb_000120_150 |
0.2956889491 |
- |
- |
hypothetical protein JCGZ_08453 [Jatropha curcas] |
| 9 |
Hb_007553_020 |
0.2976837966 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004814_010 |
0.3010486233 |
- |
- |
PREDICTED: uncharacterized protein LOC105633486 [Jatropha curcas] |
| 11 |
Hb_000042_360 |
0.3033334947 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001597_050 |
0.3057540206 |
- |
- |
- |
| 13 |
Hb_104764_020 |
0.3061916441 |
- |
- |
hypothetical protein TRIUR3_08295 [Triticum urartu] |
| 14 |
Hb_002539_100 |
0.3115922441 |
- |
- |
PREDICTED: 14-3-3-like protein GF14 iota [Jatropha curcas] |
| 15 |
Hb_000768_140 |
0.3119183579 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
tRNA (guanine-n(7)-)-methyltransferase, putative [Ricinus communis] |
| 16 |
Hb_004453_070 |
0.312703278 |
transcription factor |
TF Family: CPP |
PREDICTED: CRC domain-containing protein TSO1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_001322_070 |
0.3130446631 |
- |
- |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_128473_010 |
0.3147908611 |
- |
- |
- |
| 19 |
Hb_000261_170 |
0.3150778235 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 20 |
Hb_001804_030 |
0.318916684 |
- |
- |
kinase, putative [Ricinus communis] |