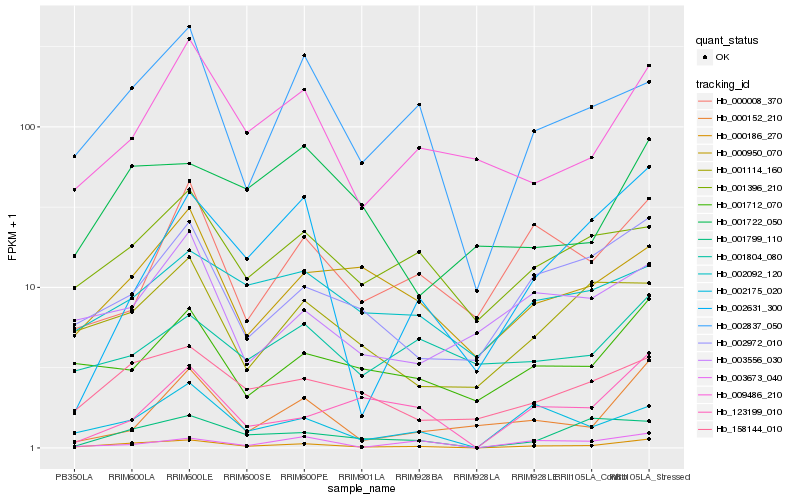
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000186_270 |
0.0 |
- |
- |
hypothetical protein JCGZ_02112 [Jatropha curcas] |
| 2 |
Hb_002175_020 |
0.1925159006 |
- |
- |
PREDICTED: uncharacterized protein LOC105642125 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002837_050 |
0.194783144 |
- |
- |
hypothetical protein POPTR_0006s00550g [Populus trichocarpa] |
| 4 |
Hb_158144_010 |
0.1986713795 |
- |
- |
- |
| 5 |
Hb_002972_010 |
0.2028096688 |
- |
- |
PREDICTED: magnesium-dependent phosphatase 1 [Jatropha curcas] |
| 6 |
Hb_001712_070 |
0.2081940699 |
- |
- |
PREDICTED: HD domain-containing protein 2 [Jatropha curcas] |
| 7 |
Hb_001799_110 |
0.2090269795 |
- |
- |
PREDICTED: glycine--tRNA ligase 2, chloroplastic/mitochondrial isoform X2 [Jatropha curcas] |
| 8 |
Hb_009486_210 |
0.2105213411 |
- |
- |
- |
| 9 |
Hb_000950_070 |
0.2212650597 |
- |
- |
PREDICTED: uncharacterized protein LOC105645259 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001396_210 |
0.2221146059 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001114_160 |
0.2221943302 |
- |
- |
PREDICTED: beta carbonic anhydrase 5, chloroplastic [Jatropha curcas] |
| 12 |
Hb_003673_040 |
0.2301702443 |
- |
- |
dynamin family protein [Populus trichocarpa] |
| 13 |
Hb_123199_010 |
0.233117992 |
- |
- |
SRK protein [Brassica cretica] |
| 14 |
Hb_002092_120 |
0.2355333901 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_002631_300 |
0.2362549642 |
- |
- |
PREDICTED: uncharacterized protein LOC105646181 [Jatropha curcas] |
| 16 |
Hb_000152_210 |
0.2378797142 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001722_050 |
0.2388163703 |
- |
- |
calcium binding family protein [Populus trichocarpa] |
| 18 |
Hb_003556_030 |
0.2417756596 |
- |
- |
PREDICTED: uncharacterized oxidoreductase C663.09c isoform X1 [Jatropha curcas] |
| 19 |
Hb_000008_370 |
0.2420221799 |
- |
- |
PREDICTED: uncharacterized protein LOC105628095 [Jatropha curcas] |
| 20 |
Hb_001804_080 |
0.2461402109 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C9-like [Jatropha curcas] |