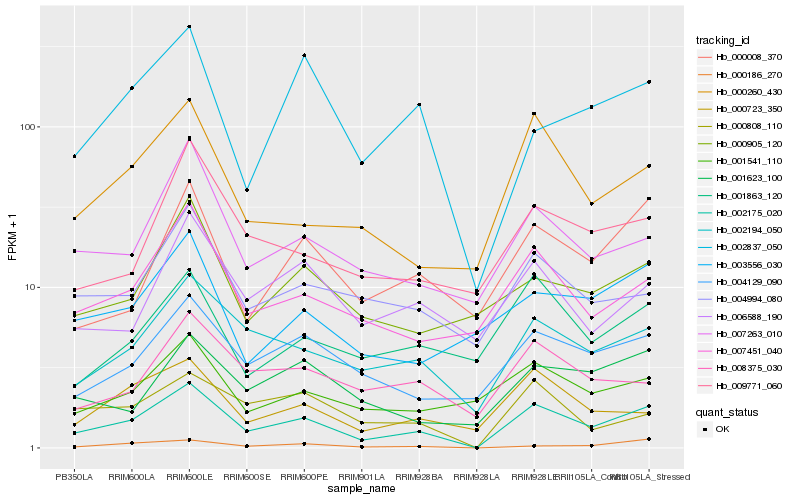
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002175_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642125 isoform X2 [Jatropha curcas] |
| 2 |
Hb_002194_050 |
0.1611459444 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Jatropha curcas] |
| 3 |
Hb_004129_090 |
0.1771479913 |
- |
- |
PREDICTED: uncharacterized protein LOC105646039 [Jatropha curcas] |
| 4 |
Hb_007451_040 |
0.17833763 |
- |
- |
PREDICTED: uncharacterized protein LOC105643411 [Jatropha curcas] |
| 5 |
Hb_000260_430 |
0.1792993079 |
- |
- |
ribosomal protein L17, putative [Ricinus communis] |
| 6 |
Hb_007263_010 |
0.1852337391 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 7 |
Hb_000808_110 |
0.1855653307 |
- |
- |
PREDICTED: uncharacterized protein LOC105647448 isoform X4 [Jatropha curcas] |
| 8 |
Hb_000723_350 |
0.1864952673 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 9 |
Hb_000008_370 |
0.1867034501 |
- |
- |
PREDICTED: uncharacterized protein LOC105628095 [Jatropha curcas] |
| 10 |
Hb_000186_270 |
0.1925159006 |
- |
- |
hypothetical protein JCGZ_02112 [Jatropha curcas] |
| 11 |
Hb_001863_120 |
0.1952603446 |
- |
- |
PREDICTED: serine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 12 |
Hb_001541_110 |
0.1953775013 |
- |
- |
PREDICTED: sufE-like protein 2, chloroplastic isoform X2 [Jatropha curcas] |
| 13 |
Hb_009771_060 |
0.1954934277 |
- |
- |
PREDICTED: 30S ribosomal protein S31, chloroplastic [Populus euphratica] |
| 14 |
Hb_004994_080 |
0.1955755312 |
- |
- |
PREDICTED: cell division protein FtsY homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_001623_100 |
0.1975671443 |
- |
- |
Hydroxyethylthiazole kinase, putative [Ricinus communis] |
| 16 |
Hb_008375_030 |
0.1979012714 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X1 [Populus euphratica] |
| 17 |
Hb_002837_050 |
0.1983488519 |
- |
- |
hypothetical protein POPTR_0006s00550g [Populus trichocarpa] |
| 18 |
Hb_006588_190 |
0.2001971159 |
- |
- |
PREDICTED: ataxin-3 homolog [Jatropha curcas] |
| 19 |
Hb_003556_030 |
0.2011255795 |
- |
- |
PREDICTED: uncharacterized oxidoreductase C663.09c isoform X1 [Jatropha curcas] |
| 20 |
Hb_000905_120 |
0.2015512421 |
- |
- |
PREDICTED: uncharacterized protein LOC105628677 [Jatropha curcas] |