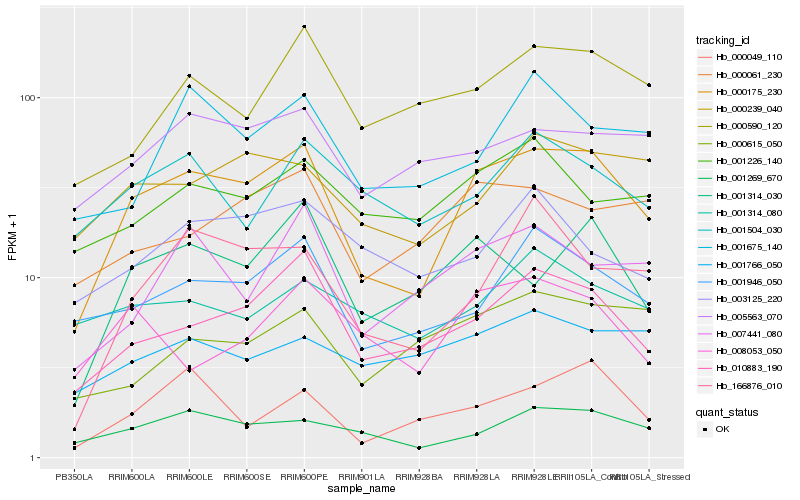
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000175_230 |
0.0 |
- |
- |
PREDICTED: protein LURP-one-related 5 [Jatropha curcas] |
| 2 |
Hb_010883_190 |
0.1377708638 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001269_670 |
0.1527033118 |
- |
- |
PREDICTED: uncharacterized protein LOC105630287 [Jatropha curcas] |
| 4 |
Hb_001946_050 |
0.1575446931 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001314_030 |
0.1588040703 |
- |
- |
PREDICTED: trifunctional UDP-glucose 4,6-dehydratase/UDP-4-keto-6-deoxy-D-glucose 3,5-epimerase/UDP-4-keto-L-rhamnose-reductase RHM1 [Jatropha curcas] |
| 6 |
Hb_166876_010 |
0.169809317 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor ids-4, putative [Ricinus communis] |
| 7 |
Hb_000239_040 |
0.1699241819 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 8 |
Hb_000061_230 |
0.1716290697 |
- |
- |
PREDICTED: putative glycosyltransferase 5 [Jatropha curcas] |
| 9 |
Hb_000590_120 |
0.1718982508 |
- |
- |
dehydroascorbate reductase, putative [Ricinus communis] |
| 10 |
Hb_007441_080 |
0.1738925657 |
- |
- |
PREDICTED: ELMO domain-containing protein C-like [Jatropha curcas] |
| 11 |
Hb_005563_070 |
0.1740214795 |
- |
- |
PREDICTED: uncharacterized protein LOC105630108 [Jatropha curcas] |
| 12 |
Hb_001675_140 |
0.1802738204 |
- |
- |
hypothetical protein L484_026216 [Morus notabilis] |
| 13 |
Hb_000049_110 |
0.1805127595 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.2 [Jatropha curcas] |
| 14 |
Hb_001226_140 |
0.1812920779 |
- |
- |
PREDICTED: protein phosphatase 2C 77-like [Jatropha curcas] |
| 15 |
Hb_001504_030 |
0.1814381657 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 6 [Jatropha curcas] |
| 16 |
Hb_008053_050 |
0.1845566303 |
- |
- |
PREDICTED: uncharacterized protein LOC105642980 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000615_050 |
0.1888278555 |
- |
- |
Potassium transporter 11 family protein [Populus trichocarpa] |
| 18 |
Hb_003125_220 |
0.1902652637 |
- |
- |
PREDICTED: CBS domain-containing protein CBSCBSPB1 [Jatropha curcas] |
| 19 |
Hb_001766_050 |
0.191291621 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29290 [Jatropha curcas] |
| 20 |
Hb_001314_080 |
0.1913690865 |
- |
- |
hypothetical protein POPTR_0018s14330g [Populus trichocarpa] |