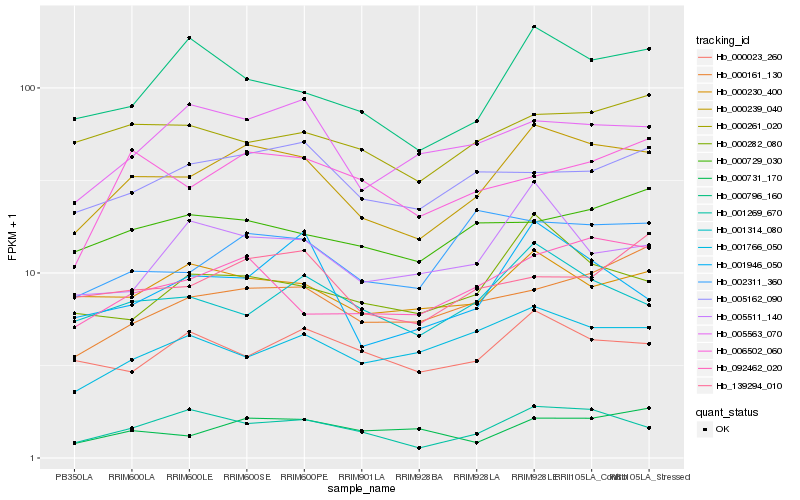
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000239_040 |
0.0 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 2 |
Hb_092462_020 |
0.1118845984 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_001269_670 |
0.1137121576 |
- |
- |
PREDICTED: uncharacterized protein LOC105630287 [Jatropha curcas] |
| 4 |
Hb_000282_080 |
0.1190693104 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000230_400 |
0.1204509902 |
- |
- |
PREDICTED: protein ABCI12, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_000796_160 |
0.1209477078 |
- |
- |
PREDICTED: nifU-like protein 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001314_080 |
0.1227196666 |
- |
- |
hypothetical protein POPTR_0018s14330g [Populus trichocarpa] |
| 8 |
Hb_005162_090 |
0.1289252635 |
- |
- |
PREDICTED: uncharacterized protein LOC105648425 [Jatropha curcas] |
| 9 |
Hb_005563_070 |
0.1296948805 |
- |
- |
PREDICTED: uncharacterized protein LOC105630108 [Jatropha curcas] |
| 10 |
Hb_001946_050 |
0.1299700873 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002311_360 |
0.1305252454 |
- |
- |
PREDICTED: uncharacterized protein LOC105114273 [Populus euphratica] |
| 12 |
Hb_001766_050 |
0.1306669371 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29290 [Jatropha curcas] |
| 13 |
Hb_005511_140 |
0.131141155 |
- |
- |
Ubiquinone biosynthesis protein coq-8, putative [Ricinus communis] |
| 14 |
Hb_000261_020 |
0.1313582638 |
- |
- |
PREDICTED: chaperone protein dnaJ 20, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000161_130 |
0.1320426214 |
- |
- |
PREDICTED: nifU-like protein 2, chloroplastic [Jatropha curcas] |
| 16 |
Hb_139294_010 |
0.1320438192 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_000731_170 |
0.1328530051 |
- |
- |
PREDICTED: phospholipase A I [Jatropha curcas] |
| 18 |
Hb_006502_060 |
0.1348416972 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000023_260 |
0.1352490017 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000729_030 |
0.1353035226 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Jatropha curcas] |