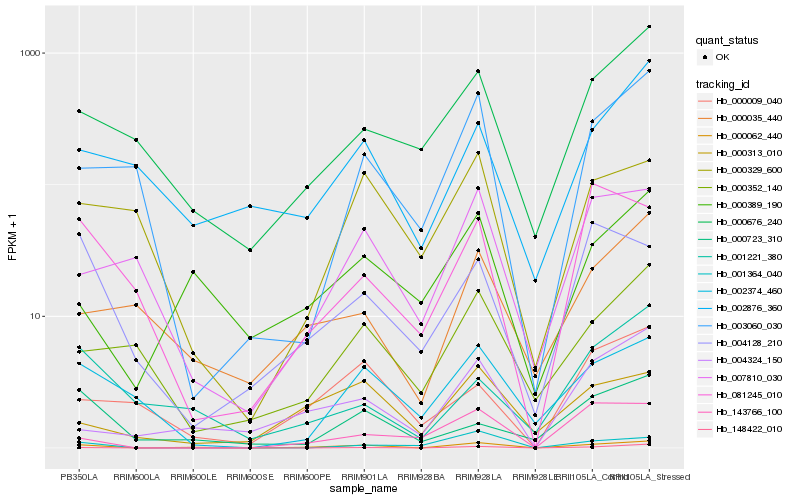
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000062_440 |
0.0 |
- |
- |
hypothetical protein JCGZ_17832 [Jatropha curcas] |
| 2 |
Hb_004324_150 |
0.260163184 |
- |
- |
- |
| 3 |
Hb_000389_190 |
0.2614217101 |
- |
- |
PREDICTED: protein PHLOEM PROTEIN 2-LIKE A1-like [Jatropha curcas] |
| 4 |
Hb_148422_010 |
0.268008677 |
- |
- |
PREDICTED: uncharacterized protein LOC104908874, partial [Beta vulgaris subsp. vulgaris] |
| 5 |
Hb_002374_460 |
0.2687356839 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000676_240 |
0.27153402 |
- |
- |
thioredoxin H-type 2 [Hevea brasiliensis] |
| 7 |
Hb_143766_100 |
0.2750635038 |
- |
- |
PREDICTED: uncharacterized protein LOC105637800 [Jatropha curcas] |
| 8 |
Hb_007810_030 |
0.2762894445 |
- |
- |
PREDICTED: uncharacterized protein LOC105647693 [Jatropha curcas] |
| 9 |
Hb_000035_440 |
0.2783866897 |
- |
- |
- |
| 10 |
Hb_000723_310 |
0.2784274375 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_000313_010 |
0.2785202463 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 12 |
Hb_002876_360 |
0.28161586 |
- |
- |
ubiquitin-conjugating enzyme E2, putative [Ricinus communis] |
| 13 |
Hb_003060_030 |
0.2818970224 |
- |
- |
conserved hypothetical protein 16 [Hevea brasiliensis] |
| 14 |
Hb_001364_040 |
0.2821419198 |
- |
- |
- |
| 15 |
Hb_001221_380 |
0.2824540163 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 30 [Jatropha curcas] |
| 16 |
Hb_004128_210 |
0.2852252061 |
- |
- |
PREDICTED: protein ALUMINUM SENSITIVE 3 [Jatropha curcas] |
| 17 |
Hb_000352_140 |
0.2856797816 |
- |
- |
- |
| 18 |
Hb_081245_010 |
0.286499887 |
- |
- |
Caffeic acid 3-O-methyltransferase [Morus notabilis] |
| 19 |
Hb_000009_040 |
0.2870016222 |
- |
- |
- |
| 20 |
Hb_000329_600 |
0.2881288345 |
- |
- |
PREDICTED: cell number regulator 6-like isoform X2 [Populus euphratica] |