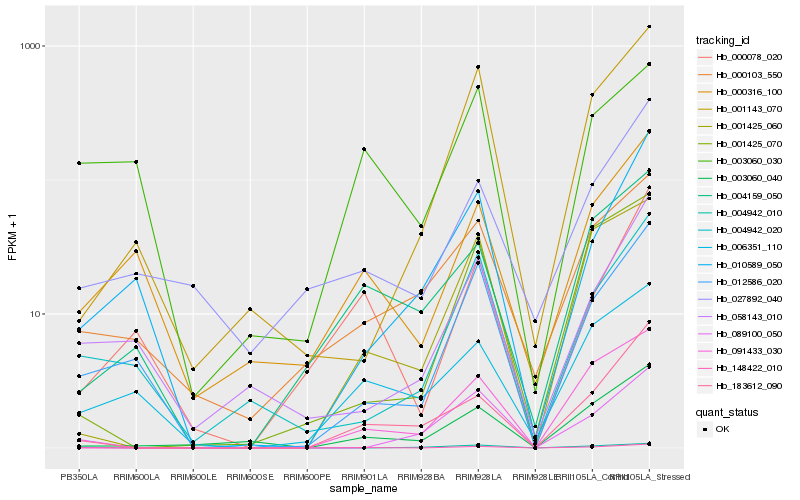
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_148422_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104908874, partial [Beta vulgaris subsp. vulgaris] |
| 2 |
Hb_183612_090 |
0.2077840948 |
- |
- |
- |
| 3 |
Hb_012586_020 |
0.2077898777 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003060_040 |
0.2294401717 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL15 [Jatropha curcas] |
| 5 |
Hb_004942_020 |
0.2311764374 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 6 |
Hb_001425_060 |
0.2335961064 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_091433_030 |
0.2336959007 |
- |
- |
- |
| 8 |
Hb_000078_020 |
0.2384655396 |
- |
- |
- |
| 9 |
Hb_001425_070 |
0.2407082833 |
- |
- |
- |
| 10 |
Hb_000316_100 |
0.2425189345 |
- |
- |
protein with unknown function [Ricinus communis] |
| 11 |
Hb_058143_010 |
0.2472202348 |
- |
- |
hypothetical protein JCGZ_01992 [Jatropha curcas] |
| 12 |
Hb_089100_050 |
0.2486216151 |
- |
- |
- |
| 13 |
Hb_006351_110 |
0.2519654904 |
- |
- |
- |
| 14 |
Hb_003060_030 |
0.2534797115 |
- |
- |
conserved hypothetical protein 16 [Hevea brasiliensis] |
| 15 |
Hb_004159_050 |
0.2543504775 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_010589_050 |
0.2580625782 |
- |
- |
- |
| 17 |
Hb_027892_040 |
0.2581772014 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |
| 18 |
Hb_000103_550 |
0.2584980265 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit gamma 2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001143_070 |
0.2592393083 |
- |
- |
Acyl-CoA N-acyltransferases (NAT) superfamily protein isoform 1 [Theobroma cacao] |
| 20 |
Hb_004942_010 |
0.2602953024 |
- |
- |
hypothetical protein AMTR_s00057p00201180 [Amborella trichopoda] |