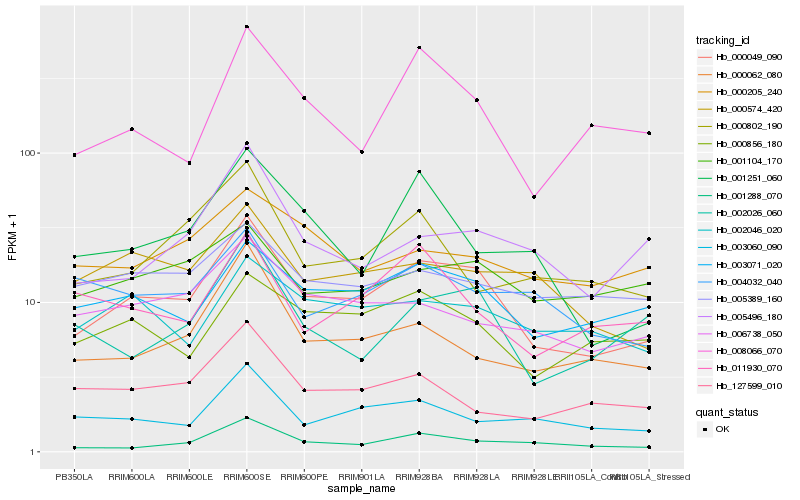
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000049_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644472 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003071_020 |
0.13557544 |
transcription factor |
TF Family: ARF |
Auxin response factor [Medicago truncatula] |
| 3 |
Hb_001288_070 |
0.1445233131 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 4 |
Hb_006738_050 |
0.1527129541 |
- |
- |
PREDICTED: uncharacterized protein LOC105634992 [Jatropha curcas] |
| 5 |
Hb_000574_420 |
0.1551829462 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like [Jatropha curcas] |
| 6 |
Hb_000856_180 |
0.1554531502 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 7 |
Hb_000062_080 |
0.1560390535 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR11-like [Jatropha curcas] |
| 8 |
Hb_003060_090 |
0.1568452298 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 9 |
Hb_008066_070 |
0.1575129715 |
transcription factor |
TF Family: ERF |
AP2/ERF super family protein [Hevea brasiliensis] |
| 10 |
Hb_002026_060 |
0.1576657902 |
- |
- |
PREDICTED: uncharacterized protein LOC105645786 [Jatropha curcas] |
| 11 |
Hb_002046_020 |
0.1604013622 |
- |
- |
PREDICTED: putative clathrin assembly protein At2g25430 [Jatropha curcas] |
| 12 |
Hb_005496_180 |
0.164445654 |
- |
- |
PREDICTED: U-box domain-containing protein 4-like [Jatropha curcas] |
| 13 |
Hb_127599_010 |
0.1667342746 |
- |
- |
hypothetical protein CISIN_1g0010462mg, partial [Citrus sinensis] |
| 14 |
Hb_001104_170 |
0.1676229946 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-2c [Jatropha curcas] |
| 15 |
Hb_004032_040 |
0.1683104679 |
- |
- |
unknown [Medicago truncatula] |
| 16 |
Hb_005389_160 |
0.1695154026 |
- |
- |
PREDICTED: casein kinase I [Jatropha curcas] |
| 17 |
Hb_011930_070 |
0.170242705 |
- |
- |
PREDICTED: probable AMP deaminase [Jatropha curcas] |
| 18 |
Hb_000802_190 |
0.1703186206 |
- |
- |
PREDICTED: peroxisomal (S)-2-hydroxy-acid oxidase [Jatropha curcas] |
| 19 |
Hb_001251_060 |
0.1706979956 |
- |
- |
aldo/keto reductase AKR [Manihot esculenta] |
| 20 |
Hb_000205_240 |
0.1716259446 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |