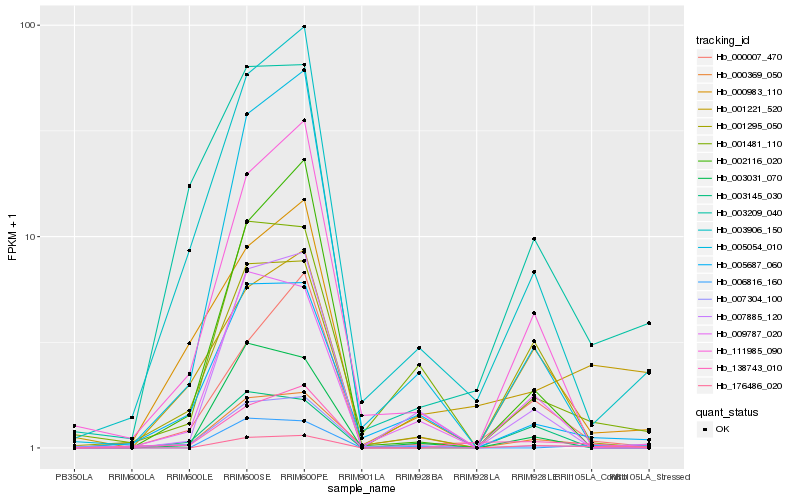
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_176486_020 |
0.0 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IX.1-like [Populus euphratica] |
| 2 |
Hb_006816_160 |
0.242928747 |
- |
- |
hypothetical protein RCOM_1434920 [Ricinus communis] |
| 3 |
Hb_138743_010 |
0.2527249692 |
- |
- |
hypothetical protein JCGZ_23770 [Jatropha curcas] |
| 4 |
Hb_001295_050 |
0.2555229462 |
- |
- |
PREDICTED: uncharacterized protein LOC101503108 isoform X1 [Cicer arietinum] |
| 5 |
Hb_000007_470 |
0.2583017464 |
- |
- |
PREDICTED: phosphate transporter PHO1 homolog 1 [Jatropha curcas] |
| 6 |
Hb_007304_100 |
0.2610816162 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g34110 [Jatropha curcas] |
| 7 |
Hb_000369_050 |
0.2636563545 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 8 |
Hb_003209_040 |
0.2674312909 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase protein 22-like [Gossypium raimondii] |
| 9 |
Hb_001481_110 |
0.268465126 |
- |
- |
S-adenosylmethionine synthetase 1 family protein [Populus trichocarpa] |
| 10 |
Hb_007885_120 |
0.2686019534 |
- |
- |
PREDICTED: BON1-associated protein 2-like [Jatropha curcas] |
| 11 |
Hb_111985_090 |
0.2690617312 |
- |
- |
hypothetical protein RCOM_1595950 [Ricinus communis] |
| 12 |
Hb_003031_070 |
0.2695152883 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 55-like [Jatropha curcas] |
| 13 |
Hb_003145_030 |
0.2711666012 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003906_150 |
0.2722351068 |
- |
- |
peptidoglycan-binding LysM domain-containing family protein, partial [Populus trichocarpa] |
| 15 |
Hb_000983_110 |
0.2741933201 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 16 |
Hb_009787_020 |
0.2748504635 |
- |
- |
PREDICTED: cytochrome P450 705A5-like [Jatropha curcas] |
| 17 |
Hb_005687_060 |
0.2751449359 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_001221_520 |
0.2757298465 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002116_020 |
0.2779246155 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 35 [Jatropha curcas] |
| 20 |
Hb_005054_010 |
0.278082793 |
- |
- |
amino acid transporter, putative [Ricinus communis] |