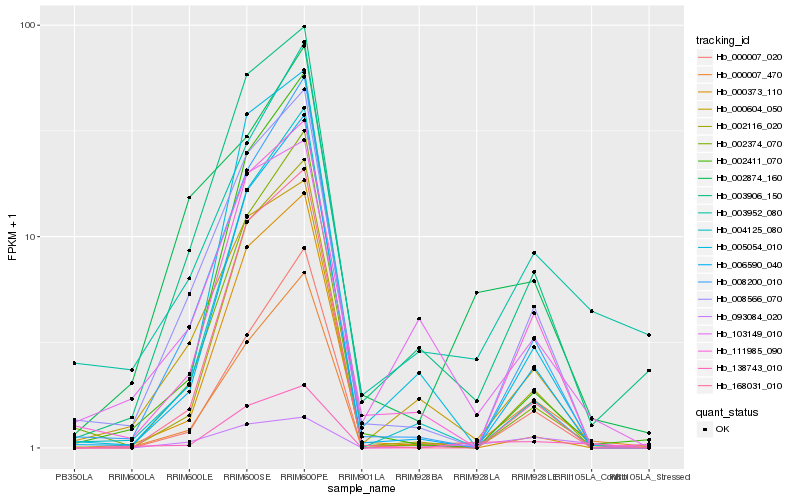
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_138743_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_23770 [Jatropha curcas] |
| 2 |
Hb_000007_470 |
0.1702316582 |
- |
- |
PREDICTED: phosphate transporter PHO1 homolog 1 [Jatropha curcas] |
| 3 |
Hb_003906_150 |
0.1952250082 |
- |
- |
peptidoglycan-binding LysM domain-containing family protein, partial [Populus trichocarpa] |
| 4 |
Hb_168031_010 |
0.2003709575 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.6 [Jatropha curcas] |
| 5 |
Hb_002116_020 |
0.2030520428 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 35 [Jatropha curcas] |
| 6 |
Hb_006590_040 |
0.2060379111 |
- |
- |
amino acid permease, putative [Ricinus communis] |
| 7 |
Hb_111985_090 |
0.2111283646 |
- |
- |
hypothetical protein RCOM_1595950 [Ricinus communis] |
| 8 |
Hb_003952_080 |
0.2139399986 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_005054_010 |
0.2141592764 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 10 |
Hb_008566_070 |
0.2145312262 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000604_050 |
0.2149384138 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 12 |
Hb_002411_070 |
0.2159442217 |
- |
- |
PREDICTED: WAT1-related protein At5g07050 [Jatropha curcas] |
| 13 |
Hb_000373_110 |
0.2187316057 |
- |
- |
Nodulin, putative [Ricinus communis] |
| 14 |
Hb_002874_160 |
0.2194896366 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_008200_010 |
0.2209788725 |
- |
- |
PREDICTED: ABC transporter G family member 14-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_093084_020 |
0.2219401716 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 3 [Populus euphratica] |
| 17 |
Hb_004125_080 |
0.2224689359 |
- |
- |
PREDICTED: dCTP pyrophosphatase 1-like [Populus euphratica] |
| 18 |
Hb_002374_070 |
0.2244433542 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 19 |
Hb_103149_010 |
0.2256942286 |
- |
- |
hypothetical protein CISIN_1g0397312mg, partial [Citrus sinensis] |
| 20 |
Hb_000007_020 |
0.2299703007 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |