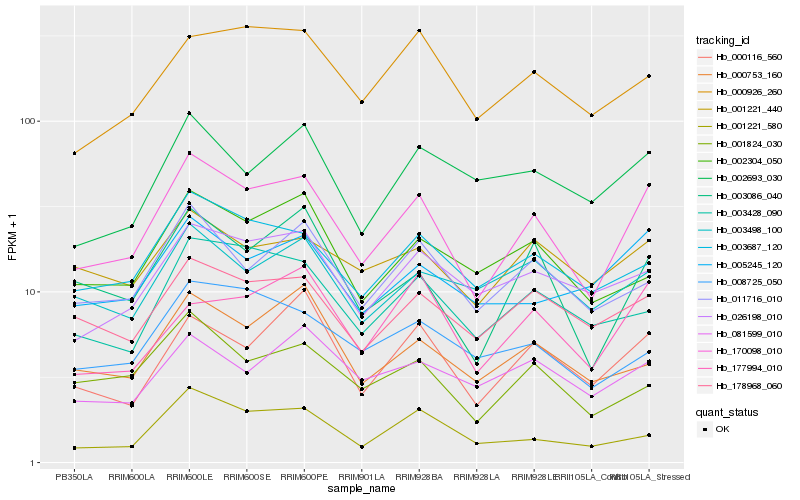
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_170098_010 |
0.0 |
- |
- |
PREDICTED: cullin-1 [Jatropha curcas] |
| 2 |
Hb_003086_040 |
0.1161174684 |
- |
- |
PREDICTED: uncharacterized protein LOC105635780 isoform X2 [Jatropha curcas] |
| 3 |
Hb_003687_120 |
0.1209230556 |
- |
- |
PREDICTED: serine/threonine-protein kinase tricorner-like [Jatropha curcas] |
| 4 |
Hb_178968_060 |
0.1239110526 |
- |
- |
PREDICTED: ubiquitin receptor RAD23b-like [Gossypium raimondii] |
| 5 |
Hb_011716_010 |
0.1243296576 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 1.2 [Jatropha curcas] |
| 6 |
Hb_000116_560 |
0.1248620007 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 [Jatropha curcas] |
| 7 |
Hb_026198_010 |
0.1274678805 |
- |
- |
PREDICTED: uncharacterized protein LOC105634369 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001824_030 |
0.1306717916 |
- |
- |
plant sec1, putative [Ricinus communis] |
| 9 |
Hb_003428_090 |
0.1312137878 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 10 |
Hb_005245_120 |
0.1313790565 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 11 |
Hb_000926_260 |
0.1350046714 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001221_440 |
0.1372302136 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 1B-1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002693_030 |
0.1375605276 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 34 kDa [Jatropha curcas] |
| 14 |
Hb_177994_010 |
0.1390643028 |
- |
- |
PREDICTED: cullin-1 [Jatropha curcas] |
| 15 |
Hb_001221_580 |
0.1402337017 |
- |
- |
PREDICTED: recQ-mediated genome instability protein 1 [Jatropha curcas] |
| 16 |
Hb_003498_100 |
0.1407393944 |
- |
- |
component of oligomeric golgi complex, putative [Ricinus communis] |
| 17 |
Hb_000753_160 |
0.1408401173 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |
| 18 |
Hb_081599_010 |
0.1409094873 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002304_050 |
0.1411263348 |
- |
- |
pyruvate kinase, putative [Ricinus communis] |
| 20 |
Hb_008725_050 |
0.1419340065 |
- |
- |
syntaxin, putative [Ricinus communis] |