| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
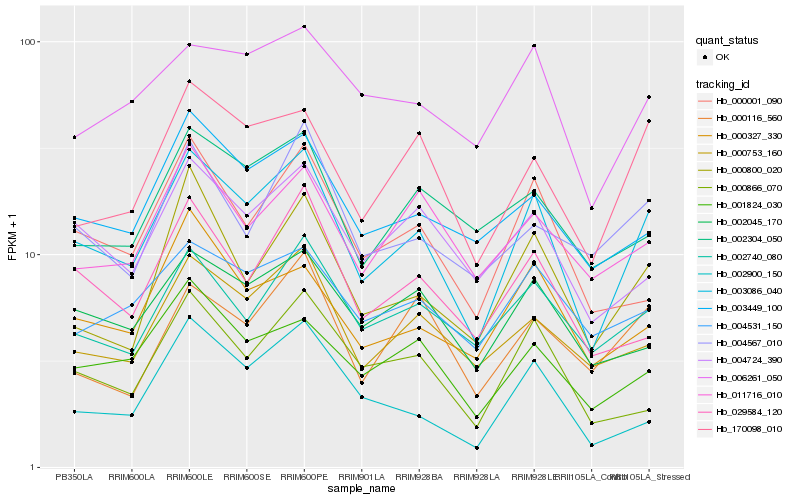
Hb_003086_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635780 isoform X2 [Jatropha curcas] |
| 2 |
Hb_170098_010 |
0.1161174684 |
- |
- |
PREDICTED: cullin-1 [Jatropha curcas] |
| 3 |
Hb_000753_160 |
0.1219883801 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |
| 4 |
Hb_001824_030 |
0.1226429061 |
- |
- |
plant sec1, putative [Ricinus communis] |
| 5 |
Hb_002740_080 |
0.1264163701 |
- |
- |
flap endonuclease-1, putative [Ricinus communis] |
| 6 |
Hb_002045_170 |
0.1268110834 |
- |
- |
interferon-induced guanylate-binding protein, putative [Ricinus communis] |
| 7 |
Hb_004724_390 |
0.1277629644 |
- |
- |
PREDICTED: nicalin-1-like [Populus euphratica] |
| 8 |
Hb_000800_020 |
0.1293988218 |
- |
- |
PREDICTED: 3-phosphoinositide-dependent protein kinase 2 [Jatropha curcas] |
| 9 |
Hb_003449_100 |
0.1301467871 |
- |
- |
PREDICTED: protein YIPF6 homolog [Jatropha curcas] |
| 10 |
Hb_000001_090 |
0.1315753416 |
- |
- |
PREDICTED: uncharacterized protein At3g58460 [Populus euphratica] |
| 11 |
Hb_000327_330 |
0.1327227197 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004531_150 |
0.1340368167 |
- |
- |
PREDICTED: peroxisome biogenesis protein 2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_006261_050 |
0.1341646407 |
transcription factor |
TF Family: C2C2-YABBY |
DNA-binding protein MNB1B, putative [Ricinus communis] |
| 14 |
Hb_029584_120 |
0.1358531683 |
- |
- |
PREDICTED: K(+) efflux antiporter 4 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000866_070 |
0.1373152503 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: SKP1-like protein 21 [Jatropha curcas] |
| 16 |
Hb_011716_010 |
0.1396528513 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 1.2 [Jatropha curcas] |
| 17 |
Hb_002304_050 |
0.1398689558 |
- |
- |
pyruvate kinase, putative [Ricinus communis] |
| 18 |
Hb_002900_150 |
0.1413118266 |
- |
- |
PREDICTED: syntaxin-81 [Jatropha curcas] |
| 19 |
Hb_000116_560 |
0.1419580526 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 [Jatropha curcas] |
| 20 |
Hb_004567_010 |
0.1429221095 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor [Jatropha curcas] |