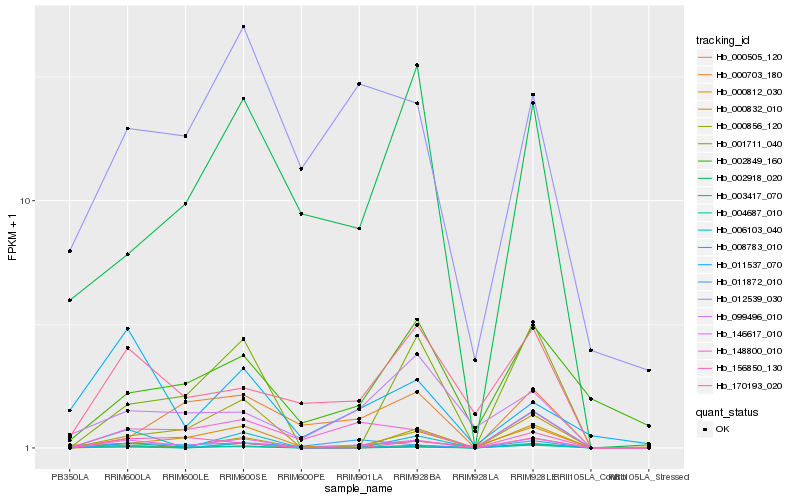
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_146617_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 2 |
Hb_148800_010 |
0.2518826778 |
- |
- |
- |
| 3 |
Hb_001711_040 |
0.2705050036 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004687_010 |
0.2910613672 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 5 |
Hb_170193_020 |
0.2953871099 |
- |
- |
- |
| 6 |
Hb_011872_010 |
0.2977547912 |
- |
- |
- |
| 7 |
Hb_000856_120 |
0.3000557064 |
transcription factor |
TF Family: LOB |
PREDICTED: myb-like protein M [Pyrus x bretschneideri] |
| 8 |
Hb_006103_040 |
0.302812591 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 9 |
Hb_008783_010 |
0.3109996957 |
- |
- |
- |
| 10 |
Hb_002918_020 |
0.313732339 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.4 isoform X1 [Jatropha curcas] |
| 11 |
Hb_012539_030 |
0.3173041882 |
- |
- |
PREDICTED: uncharacterized protein LOC105638247 [Jatropha curcas] |
| 12 |
Hb_000832_010 |
0.3177353502 |
- |
- |
hypothetical protein VITISV_005044 [Vitis vinifera] |
| 13 |
Hb_011537_070 |
0.3239115381 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Vitis vinifera] |
| 14 |
Hb_002849_160 |
0.3306638586 |
- |
- |
PREDICTED: uncharacterized protein LOC105642953 isoform X2 [Jatropha curcas] |
| 15 |
Hb_156850_130 |
0.3326733893 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_099496_010 |
0.333685585 |
- |
- |
PREDICTED: uncharacterized protein LOC105138000 [Populus euphratica] |
| 17 |
Hb_003417_070 |
0.3342896587 |
- |
- |
hypothetical protein MIMGU_mgv1a018902mg [Erythranthe guttata] |
| 18 |
Hb_000812_030 |
0.3348906054 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 19 |
Hb_000505_120 |
0.3367920491 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 20 |
Hb_000703_180 |
0.3370423513 |
- |
- |
PREDICTED: uncharacterized protein LOC104100193 [Nicotiana tomentosiformis] |