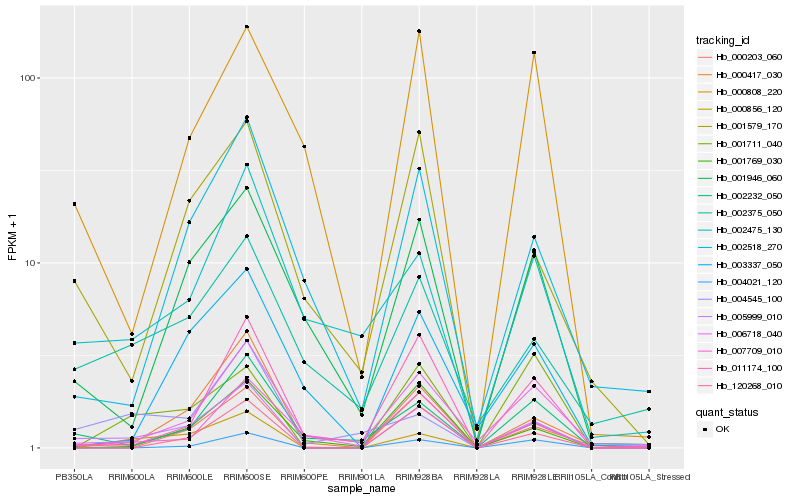
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005999_010 |
0.0 |
- |
- |
Sugar transporter ERD6-like 5 [Morus notabilis] |
| 2 |
Hb_000856_120 |
0.2186675079 |
transcription factor |
TF Family: LOB |
PREDICTED: myb-like protein M [Pyrus x bretschneideri] |
| 3 |
Hb_002232_050 |
0.2208814057 |
- |
- |
PREDICTED: uncharacterized protein LOC104214925 [Nicotiana sylvestris] |
| 4 |
Hb_000417_030 |
0.222291187 |
transcription factor |
TF Family: G2-like |
PREDICTED: putative two-component response regulator ARR19 [Jatropha curcas] |
| 5 |
Hb_001579_170 |
0.2315998928 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_006718_040 |
0.2337185652 |
- |
- |
PREDICTED: starch synthase 3, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_001946_060 |
0.2374854342 |
- |
- |
PREDICTED: TBC1 domain family member 13-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_120268_010 |
0.242565165 |
- |
- |
PREDICTED: chitin elicitor receptor kinase 1-like isoform X3 [Populus euphratica] |
| 9 |
Hb_007709_010 |
0.245711225 |
- |
- |
- |
| 10 |
Hb_002475_130 |
0.2496942496 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 4 [Jatropha curcas] |
| 11 |
Hb_004021_120 |
0.2514429798 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 12 |
Hb_011174_100 |
0.2529812776 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 13 |
Hb_002518_270 |
0.2531225204 |
- |
- |
glycogen phosphorylase, putative [Ricinus communis] |
| 14 |
Hb_002375_050 |
0.2531309191 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 15 |
Hb_004545_100 |
0.2568729222 |
- |
- |
PREDICTED: protein BONZAI 3 [Jatropha curcas] |
| 16 |
Hb_000808_220 |
0.2636841186 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump 1-like [Jatropha curcas] |
| 17 |
Hb_001769_030 |
0.2637184123 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Jatropha curcas] |
| 18 |
Hb_003337_050 |
0.2646036125 |
- |
- |
PREDICTED: probable protein phosphatase 2C 78 [Jatropha curcas] |
| 19 |
Hb_000203_060 |
0.2650727254 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0004s05660g [Populus trichocarpa] |
| 20 |
Hb_001711_040 |
0.2662449027 |
- |
- |
conserved hypothetical protein [Ricinus communis] |