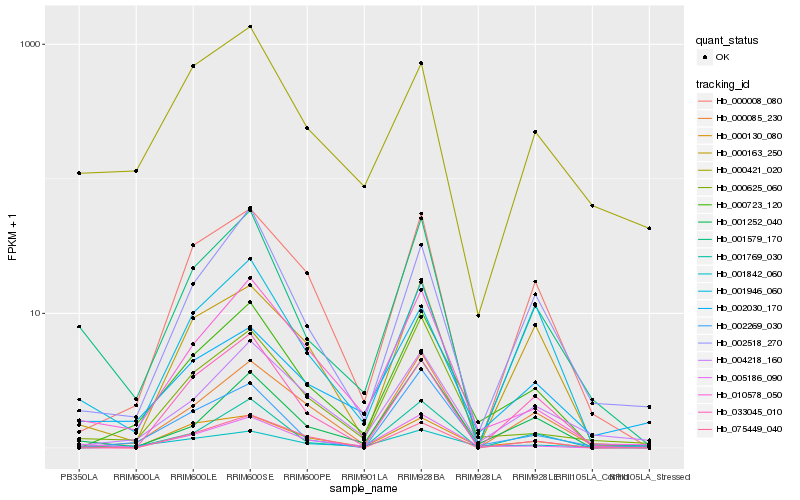
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001579_170 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_002518_270 |
0.1663554275 |
- |
- |
glycogen phosphorylase, putative [Ricinus communis] |
| 3 |
Hb_001946_060 |
0.1667145629 |
- |
- |
PREDICTED: TBC1 domain family member 13-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000421_020 |
0.1799716974 |
- |
- |
ascorbate peroxidase [Hevea brasiliensis] |
| 5 |
Hb_005186_090 |
0.1832658575 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_010578_050 |
0.1849912668 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001769_030 |
0.1859175477 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Jatropha curcas] |
| 8 |
Hb_000008_080 |
0.190541971 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002030_170 |
0.1913133802 |
- |
- |
PREDICTED: calcium-transporting ATPase 1-like [Jatropha curcas] |
| 10 |
Hb_000163_250 |
0.1924802196 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_001252_040 |
0.1935711375 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |
| 12 |
Hb_000085_230 |
0.1944813629 |
- |
- |
PREDICTED: probable long-chain-alcohol O-fatty-acyltransferase 5 [Jatropha curcas] |
| 13 |
Hb_000625_060 |
0.1964030008 |
- |
- |
Protein P21, putative [Ricinus communis] |
| 14 |
Hb_000723_120 |
0.1964846168 |
- |
- |
PREDICTED: UDP-glycosyltransferase 86A1 [Jatropha curcas] |
| 15 |
Hb_002269_030 |
0.1971273904 |
- |
- |
hypothetical protein CISIN_1g018400mg [Citrus sinensis] |
| 16 |
Hb_004218_160 |
0.2001616782 |
- |
- |
NBS-LRR type disease resistance protein [Medicago truncatula] |
| 17 |
Hb_001842_060 |
0.2006138478 |
desease resistance |
Gene Name: ABC_trans_N |
hypothetical protein JCGZ_10908 [Jatropha curcas] |
| 18 |
Hb_075449_040 |
0.2011241833 |
- |
- |
PREDICTED: meiotic recombination protein SPO11-1 [Jatropha curcas] |
| 19 |
Hb_033045_010 |
0.2032813358 |
- |
- |
copine, putative [Ricinus communis] |
| 20 |
Hb_000130_080 |
0.2041961941 |
- |
- |
unnamed protein product [Vitis vinifera] |