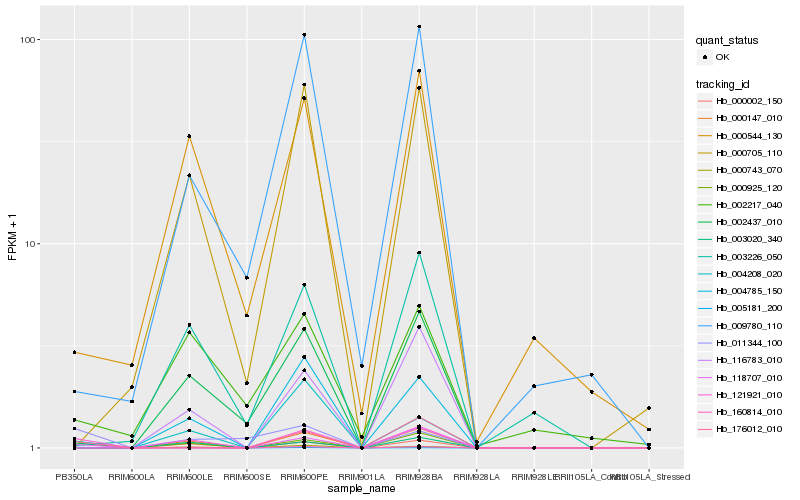
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_121921_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_004208_020 |
0.2793427957 |
- |
- |
phosphoglycerate kinase, putative [Ricinus communis] |
| 3 |
Hb_002437_010 |
0.2882293737 |
- |
- |
PREDICTED: uncharacterized protein LOC105123193 [Populus euphratica] |
| 4 |
Hb_000147_010 |
0.2896960325 |
- |
- |
PREDICTED: UDP-glycosyltransferase 87A1-like [Jatropha curcas] |
| 5 |
Hb_011344_100 |
0.2921895291 |
- |
- |
- |
| 6 |
Hb_005181_200 |
0.2924558304 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein RGL1-like [Jatropha curcas] |
| 7 |
Hb_000544_130 |
0.2943974476 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 8 |
Hb_176012_010 |
0.295284993 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_160814_010 |
0.2964843721 |
- |
- |
PREDICTED: probable sulfate transporter 3.5 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000002_150 |
0.2990787455 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000925_120 |
0.301125123 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |
| 12 |
Hb_118707_010 |
0.302331362 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 13 |
Hb_000705_110 |
0.3065506564 |
- |
- |
hypothetical protein POPTR_0005s09220g [Populus trichocarpa] |
| 14 |
Hb_003020_340 |
0.3095389622 |
- |
- |
PREDICTED: uncharacterized protein LOC105631327 [Jatropha curcas] |
| 15 |
Hb_116783_010 |
0.311807664 |
- |
- |
hypothetical protein POPTR_0011s16190g [Populus trichocarpa] |
| 16 |
Hb_002217_040 |
0.3123386699 |
- |
- |
PREDICTED: boron transporter 1-like [Jatropha curcas] |
| 17 |
Hb_004785_150 |
0.3125668689 |
- |
- |
hypothetical protein JCGZ_25207 [Jatropha curcas] |
| 18 |
Hb_000743_070 |
0.3163848418 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_21060 [Jatropha curcas] |
| 19 |
Hb_009780_110 |
0.3166480537 |
- |
- |
PREDICTED: protein RSI-1 [Cicer arietinum] |
| 20 |
Hb_003226_050 |
0.3203144445 |
- |
- |
Pectinesterase-3 precursor, putative [Ricinus communis] |