| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
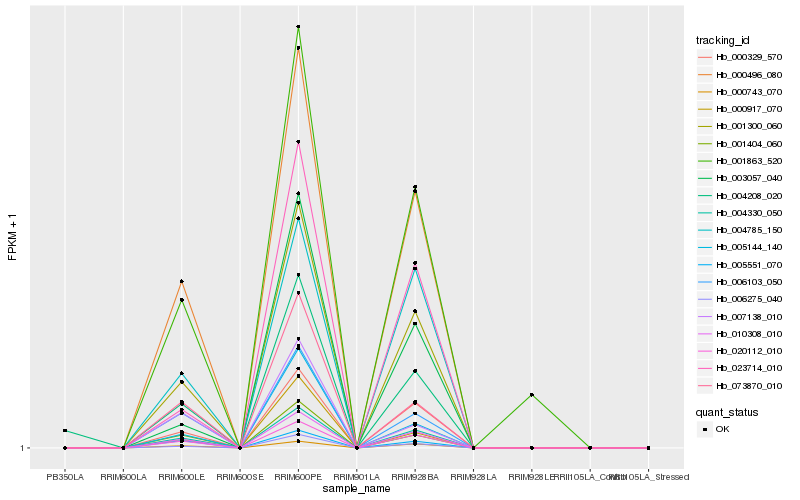
Hb_004208_020 |
0.0 |
- |
- |
phosphoglycerate kinase, putative [Ricinus communis] |
| 2 |
Hb_004330_050 |
0.1523400058 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001300_060 |
0.1523552033 |
- |
- |
PREDICTED: monothiol glutaredoxin-S6-like [Jatropha curcas] |
| 4 |
Hb_010308_010 |
0.1527838955 |
- |
- |
hypothetical protein JCGZ_22818 [Jatropha curcas] |
| 5 |
Hb_000496_080 |
0.1530779895 |
- |
- |
Proteinase inhibitor, putative [Ricinus communis] |
| 6 |
Hb_020112_010 |
0.158452109 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 7 |
Hb_006275_040 |
0.1592703951 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 8 |
Hb_000329_570 |
0.165719851 |
- |
- |
- |
| 9 |
Hb_001404_060 |
0.1665936594 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_006103_050 |
0.1687649017 |
- |
- |
- |
| 11 |
Hb_073870_010 |
0.1691723608 |
- |
- |
hypothetical protein RCOM_0725510 [Ricinus communis] |
| 12 |
Hb_005144_140 |
0.1787584675 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000743_070 |
0.1839123753 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_21060 [Jatropha curcas] |
| 14 |
Hb_000917_070 |
0.1886705157 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005551_070 |
0.1890494333 |
- |
- |
PREDICTED: uncharacterized protein LOC105635822 isoform X1 [Jatropha curcas] |
| 16 |
Hb_004785_150 |
0.1923571585 |
- |
- |
hypothetical protein JCGZ_25207 [Jatropha curcas] |
| 17 |
Hb_001863_520 |
0.193160306 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_023714_010 |
0.1965501122 |
- |
- |
hypothetical protein B456_007G377100 [Gossypium raimondii] |
| 19 |
Hb_007138_010 |
0.1990159188 |
- |
- |
PREDICTED: putative laccase-9 [Jatropha curcas] |
| 20 |
Hb_003057_040 |
0.1996924079 |
- |
- |
PREDICTED: salt stress-induced hydrophobic peptide ESI3 [Vitis vinifera] |