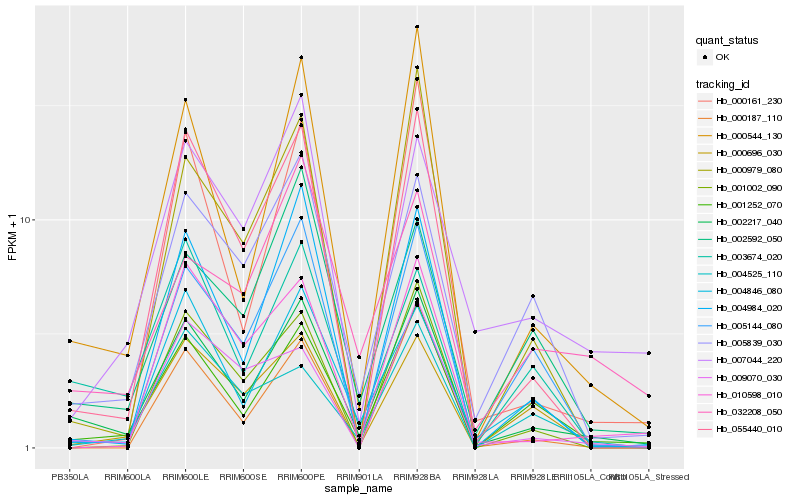
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002217_040 |
0.0 |
- |
- |
PREDICTED: boron transporter 1-like [Jatropha curcas] |
| 2 |
Hb_000544_130 |
0.1255278826 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 3 |
Hb_001252_070 |
0.1277413033 |
- |
- |
hypothetical protein JCGZ_01385 [Jatropha curcas] |
| 4 |
Hb_005839_030 |
0.1586896792 |
- |
- |
PREDICTED: potassium transporter 2 [Jatropha curcas] |
| 5 |
Hb_055440_010 |
0.15922098 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 6 |
Hb_003674_020 |
0.165314922 |
- |
- |
PREDICTED: DNA replication licensing factor MCM6 [Jatropha curcas] |
| 7 |
Hb_002592_050 |
0.1723410947 |
- |
- |
PREDICTED: LON peptidase N-terminal domain and RING finger protein 1-like [Jatropha curcas] |
| 8 |
Hb_004984_020 |
0.1740883761 |
- |
- |
PREDICTED: uncharacterized protein LOC105647814 [Jatropha curcas] |
| 9 |
Hb_004846_080 |
0.1759086806 |
- |
- |
DNA replication licensing factor MCM3, putative [Ricinus communis] |
| 10 |
Hb_005144_080 |
0.1784638141 |
- |
- |
Serine carboxypeptidase, putative [Ricinus communis] |
| 11 |
Hb_000696_030 |
0.1831325138 |
- |
- |
PREDICTED: uncharacterized protein LOC105647085 isoform X1 [Jatropha curcas] |
| 12 |
Hb_032208_050 |
0.1864741972 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 4 isoform X3 [Jatropha curcas] |
| 13 |
Hb_000979_080 |
0.188272867 |
- |
- |
PREDICTED: 21 kDa protein-like [Jatropha curcas] |
| 14 |
Hb_007044_220 |
0.1893126926 |
- |
- |
Allene oxide cyclase 4, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_000161_230 |
0.1912605409 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 16 |
Hb_009070_030 |
0.1932885955 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 10 [Jatropha curcas] |
| 17 |
Hb_001002_090 |
0.194490119 |
- |
- |
PREDICTED: receptor protein kinase-like protein At4g34220 [Jatropha curcas] |
| 18 |
Hb_010598_010 |
0.1946924761 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |
| 19 |
Hb_000187_110 |
0.1966717475 |
- |
- |
PREDICTED: serine carboxypeptidase-like 7 [Jatropha curcas] |
| 20 |
Hb_004525_110 |
0.2010666819 |
- |
- |
hypothetical protein POPTR_0007s12850g [Populus trichocarpa] |