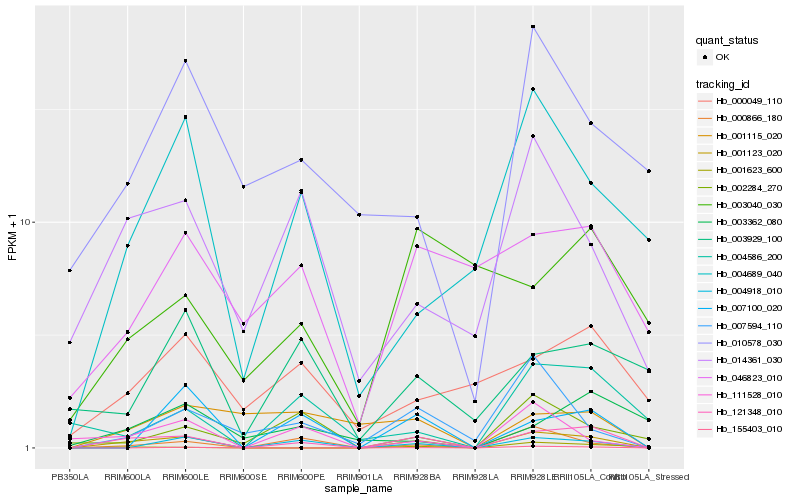
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_121348_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_001123_020 |
0.2220543023 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein CLAVATA2 [Jatropha curcas] |
| 3 |
Hb_004918_010 |
0.3378881074 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 4 |
Hb_000866_180 |
0.3530200256 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Populus euphratica] |
| 5 |
Hb_007100_020 |
0.3535811693 |
- |
- |
hypothetical protein POPTR_0473s00220g [Populus trichocarpa] |
| 6 |
Hb_155403_010 |
0.3674540177 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 7 |
Hb_014361_030 |
0.3703939343 |
- |
- |
hypothetical protein POPTR_0005s18860g [Populus trichocarpa] |
| 8 |
Hb_001623_600 |
0.3745016673 |
- |
- |
PREDICTED: uncharacterized protein LOC104748784 [Camelina sativa] |
| 9 |
Hb_004586_200 |
0.377299792 |
- |
- |
PREDICTED: cyclin-D3-1-like [Jatropha curcas] |
| 10 |
Hb_046823_010 |
0.378162687 |
- |
- |
hypothetical protein CICLE_v100021301mg, partial [Citrus clementina] |
| 11 |
Hb_000049_110 |
0.3805540677 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.2 [Jatropha curcas] |
| 12 |
Hb_004689_040 |
0.3856133187 |
- |
- |
PREDICTED: uncharacterized protein LOC105648084 [Jatropha curcas] |
| 13 |
Hb_007594_110 |
0.3911325829 |
- |
- |
fructokinase [Manihot esculenta] |
| 14 |
Hb_001115_020 |
0.3960649646 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 2-like [Populus euphratica] |
| 15 |
Hb_111528_010 |
0.396332417 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 2-like [Jatropha curcas] |
| 16 |
Hb_002284_270 |
0.3979064885 |
transcription factor |
TF Family: HB |
DNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_003362_080 |
0.4006072414 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 18 |
Hb_003929_100 |
0.4030182853 |
transcription factor |
TF Family: M-type |
PREDICTED: agamous-like MADS-box protein AGL61 [Populus euphratica] |
| 19 |
Hb_003040_030 |
0.4037163419 |
- |
- |
PREDICTED: NADP-dependent malic enzyme isoform X1 [Jatropha curcas] |
| 20 |
Hb_010578_030 |
0.4064293234 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATA, chloroplastic [Jatropha curcas] |