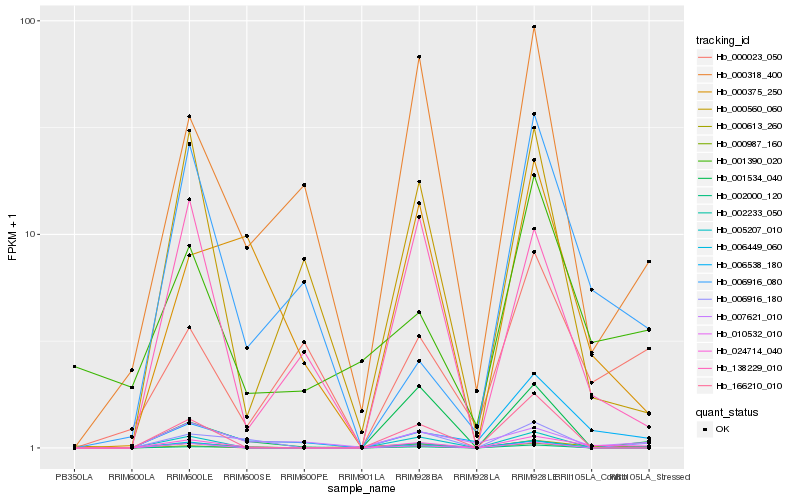
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_024714_040 |
0.0 |
- |
- |
PREDICTED: DNA mismatch repair protein PMS1 [Jatropha curcas] |
| 2 |
Hb_006538_180 |
0.257172518 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger family protein [Populus trichocarpa] |
| 3 |
Hb_006916_180 |
0.2611331039 |
- |
- |
Mitochondrial carnitine/acylcarnitine carrier protein, putative [Ricinus communis] |
| 4 |
Hb_000613_260 |
0.2716584324 |
- |
- |
hypothetical protein VITISV_043487 [Vitis vinifera] |
| 5 |
Hb_001390_020 |
0.2863893908 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_138229_010 |
0.2988048532 |
- |
- |
hypothetical protein JCGZ_05736 [Jatropha curcas] |
| 7 |
Hb_000987_160 |
0.299500595 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 8 |
Hb_002000_120 |
0.3019738783 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 9 |
Hb_006449_060 |
0.304073169 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 10 |
Hb_000318_400 |
0.304323231 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_010532_010 |
0.3043599278 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 isoform X1 [Populus euphratica] |
| 12 |
Hb_001534_040 |
0.3053227906 |
- |
- |
hypothetical protein JCGZ_08436 [Jatropha curcas] |
| 13 |
Hb_006916_080 |
0.3055145429 |
- |
- |
PREDICTED: uncharacterized protein LOC105646751 [Jatropha curcas] |
| 14 |
Hb_002233_050 |
0.305557278 |
- |
- |
PREDICTED: uncharacterized protein LOC104879391 [Vitis vinifera] |
| 15 |
Hb_007621_010 |
0.306171967 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000023_050 |
0.3070223872 |
- |
- |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 17 |
Hb_005207_010 |
0.3080910711 |
- |
- |
PREDICTED: zinc finger Ran-binding domain-containing protein 2-like [Jatropha curcas] |
| 18 |
Hb_166210_010 |
0.3086234376 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000560_060 |
0.3110160821 |
- |
- |
PREDICTED: endochitinase PR4-like [Jatropha curcas] |
| 20 |
Hb_000375_250 |
0.3124312448 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |