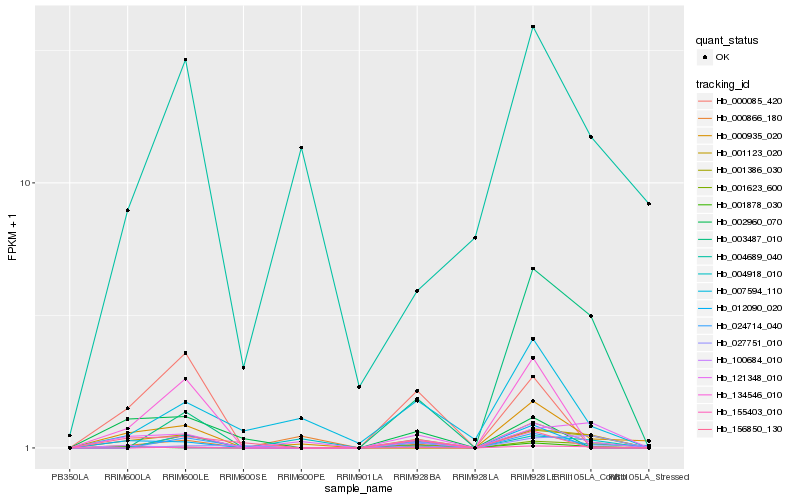
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001123_020 |
0.0 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein CLAVATA2 [Jatropha curcas] |
| 2 |
Hb_121348_010 |
0.2220543023 |
- |
- |
- |
| 3 |
Hb_155403_010 |
0.2757632257 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 4 |
Hb_001623_600 |
0.3300861522 |
- |
- |
PREDICTED: uncharacterized protein LOC104748784 [Camelina sativa] |
| 5 |
Hb_000085_420 |
0.3629777824 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_024714_040 |
0.3636145746 |
- |
- |
PREDICTED: DNA mismatch repair protein PMS1 [Jatropha curcas] |
| 7 |
Hb_003487_010 |
0.3641745636 |
- |
- |
- |
| 8 |
Hb_001878_030 |
0.3742139177 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105126050 [Populus euphratica] |
| 9 |
Hb_001386_030 |
0.3743898866 |
- |
- |
PREDICTED: peroxidase 55 [Jatropha curcas] |
| 10 |
Hb_012090_020 |
0.3764634235 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 [Jatropha curcas] |
| 11 |
Hb_007594_110 |
0.3842601178 |
- |
- |
fructokinase [Manihot esculenta] |
| 12 |
Hb_000935_020 |
0.3908870339 |
- |
- |
putative non-LTR retroelement reverse transcriptase [Arabidopsis thaliana] |
| 13 |
Hb_000866_180 |
0.3994331593 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Populus euphratica] |
| 14 |
Hb_004918_010 |
0.4022022804 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_156850_130 |
0.403385845 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_027751_010 |
0.4035545071 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 17 |
Hb_004689_040 |
0.4041534941 |
- |
- |
PREDICTED: uncharacterized protein LOC105648084 [Jatropha curcas] |
| 18 |
Hb_002960_070 |
0.4059402976 |
- |
- |
- |
| 19 |
Hb_134546_010 |
0.4063061489 |
- |
- |
PREDICTED: uncharacterized protein LOC105107372 [Populus euphratica] |
| 20 |
Hb_100684_010 |
0.4095196374 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |