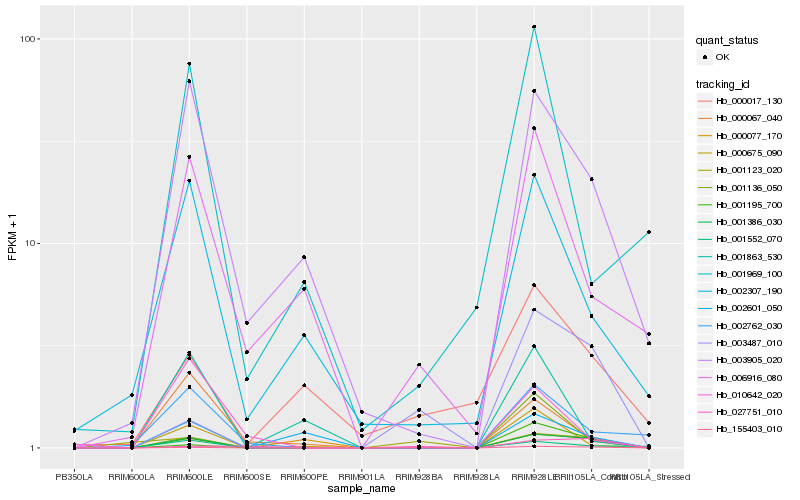
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001386_030 |
0.0 |
- |
- |
PREDICTED: peroxidase 55 [Jatropha curcas] |
| 2 |
Hb_155403_010 |
0.2296619962 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 3 |
Hb_003905_020 |
0.2735142977 |
- |
- |
PREDICTED: uncharacterized protein LOC105131878 isoform X1 [Populus euphratica] |
| 4 |
Hb_001195_700 |
0.2753320294 |
- |
- |
- |
| 5 |
Hb_001552_070 |
0.2881272418 |
- |
- |
EXS family protein [Populus trichocarpa] |
| 6 |
Hb_002762_030 |
0.3107051743 |
- |
- |
hypothetical protein POPTR_0019s12370g, partial [Populus trichocarpa] |
| 7 |
Hb_000675_090 |
0.3275157068 |
- |
- |
hypothetical protein SORBIDRAFT_03g028570 [Sorghum bicolor] |
| 8 |
Hb_000077_170 |
0.3295214403 |
- |
- |
PREDICTED: GPI-anchored protein LORELEI-like [Jatropha curcas] |
| 9 |
Hb_003487_010 |
0.334819505 |
- |
- |
- |
| 10 |
Hb_002307_190 |
0.3351530392 |
- |
- |
PREDICTED: putative ion channel POLLUX-like 2 [Jatropha curcas] |
| 11 |
Hb_002601_050 |
0.3357047088 |
- |
- |
Boron transporter, putative [Ricinus communis] |
| 12 |
Hb_001136_050 |
0.3417487587 |
- |
- |
- |
| 13 |
Hb_006916_080 |
0.3643620239 |
- |
- |
PREDICTED: uncharacterized protein LOC105646751 [Jatropha curcas] |
| 14 |
Hb_000067_040 |
0.3699251132 |
- |
- |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 15 |
Hb_001123_020 |
0.3743898866 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein CLAVATA2 [Jatropha curcas] |
| 16 |
Hb_027751_010 |
0.3764357256 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 17 |
Hb_000017_130 |
0.380047634 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog MMK2-like [Jatropha curcas] |
| 18 |
Hb_001969_100 |
0.3857003403 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 19 |
Hb_001863_530 |
0.3888912574 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_010642_020 |
0.3889842625 |
- |
- |
NDH-dependent cyclic electron flow 1 isoform 2 [Theobroma cacao] |