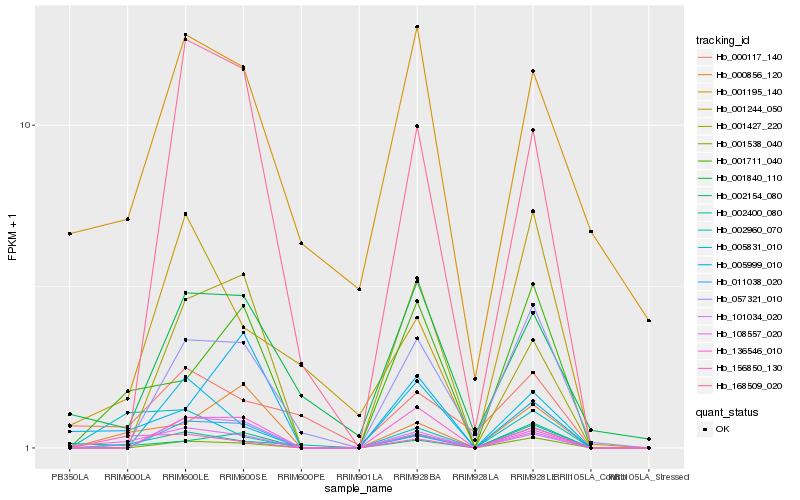
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_108557_020 |
0.0 |
transcription factor |
TF Family: B3 |
Zeamatin precursor, putative [Ricinus communis] |
| 2 |
Hb_156850_130 |
0.2358054563 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002400_080 |
0.2562440989 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 4 |
Hb_001711_040 |
0.2587405491 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002960_070 |
0.2627500817 |
- |
- |
- |
| 6 |
Hb_001840_110 |
0.2652007596 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 7 |
Hb_005831_010 |
0.2758487339 |
- |
- |
hypothetical protein JCGZ_04003 [Jatropha curcas] |
| 8 |
Hb_001195_140 |
0.2770619165 |
- |
- |
PREDICTED: U-box domain-containing protein 5-like [Jatropha curcas] |
| 9 |
Hb_000117_140 |
0.2771562217 |
- |
- |
hypothetical protein CISIN_1g042884mg, partial [Citrus sinensis] |
| 10 |
Hb_000856_120 |
0.2781522825 |
transcription factor |
TF Family: LOB |
PREDICTED: myb-like protein M [Pyrus x bretschneideri] |
| 11 |
Hb_011038_020 |
0.2785523442 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002154_080 |
0.2790576192 |
- |
- |
hypothetical protein MIMGU_mgv1a018017mg, partial [Erythranthe guttata] |
| 13 |
Hb_136546_010 |
0.2809854399 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1, partial [Populus euphratica] |
| 14 |
Hb_057321_010 |
0.2823159822 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 15 |
Hb_001427_220 |
0.2854400351 |
- |
- |
hypothetical protein CISIN_1g033375mg [Citrus sinensis] |
| 16 |
Hb_001244_050 |
0.2860383317 |
- |
- |
PREDICTED: uncharacterized protein LOC105643334 [Jatropha curcas] |
| 17 |
Hb_168509_020 |
0.2867752354 |
- |
- |
macrophage migration inhibitory factor family protein [Populus trichocarpa] |
| 18 |
Hb_005999_010 |
0.2889339232 |
- |
- |
Sugar transporter ERD6-like 5 [Morus notabilis] |
| 19 |
Hb_101034_020 |
0.2894420612 |
- |
- |
hypothetical protein JCGZ_13470 [Jatropha curcas] |
| 20 |
Hb_001538_040 |
0.2954148233 |
- |
- |
PREDICTED: uncharacterized protein LOC104228304 [Nicotiana sylvestris] |