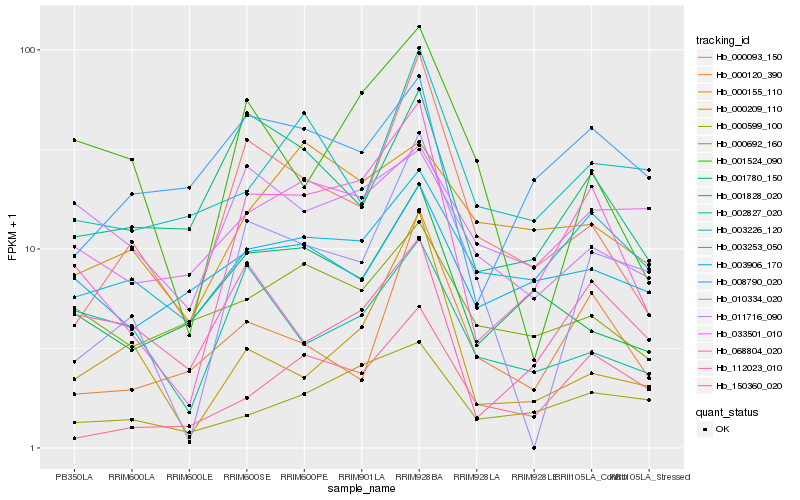
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_068804_020 |
0.0 |
- |
- |
hypothetical protein AMTR_s00046p00231970 [Amborella trichopoda] |
| 2 |
Hb_150360_020 |
0.2083268204 |
- |
- |
PREDICTED: N-alpha-acetyltransferase 11 [Jatropha curcas] |
| 3 |
Hb_000093_150 |
0.2120216654 |
- |
- |
6-phosphogluconolactonase, putative [Ricinus communis] |
| 4 |
Hb_000599_100 |
0.2157389949 |
- |
- |
hypothetical protein B456_001G254000 [Gossypium raimondii] |
| 5 |
Hb_001828_020 |
0.2198301303 |
- |
- |
hypothetical protein CISIN_1g010132mg [Citrus sinensis] |
| 6 |
Hb_001780_150 |
0.221681644 |
- |
- |
ceramidase, putative [Ricinus communis] |
| 7 |
Hb_003253_050 |
0.2289905443 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 3 [Jatropha curcas] |
| 8 |
Hb_112023_010 |
0.2355149758 |
- |
- |
hypothetical protein EUGRSUZ_C03826 [Eucalyptus grandis] |
| 9 |
Hb_010334_020 |
0.2362009124 |
- |
- |
PREDICTED: inactive protein RESTRICTED TEV MOVEMENT 1-like [Sesamum indicum] |
| 10 |
Hb_008790_020 |
0.2395919407 |
- |
- |
PREDICTED: uncharacterized protein LOC105638122 [Jatropha curcas] |
| 11 |
Hb_000120_390 |
0.2411669486 |
- |
- |
PREDICTED: uncharacterized transporter YBR287W-like [Jatropha curcas] |
| 12 |
Hb_000209_110 |
0.2427981218 |
- |
- |
PREDICTED: receptor-like protein kinase BRI1-like 3 [Jatropha curcas] |
| 13 |
Hb_001524_090 |
0.2439546133 |
- |
- |
PREDICTED: uncharacterized protein LOC105113929 [Populus euphratica] |
| 14 |
Hb_000692_160 |
0.2452740872 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |
| 15 |
Hb_011716_090 |
0.2495288727 |
transcription factor |
TF Family: HRT |
putative transciption factor family protein [Populus trichocarpa] |
| 16 |
Hb_003226_120 |
0.2500266621 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003906_170 |
0.2508456469 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_033501_010 |
0.2509037974 |
- |
- |
hypothetical protein JCGZ_21381 [Jatropha curcas] |
| 19 |
Hb_002827_020 |
0.2521504879 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 5-like [Jatropha curcas] |
| 20 |
Hb_000155_110 |
0.254603019 |
- |
- |
Auxin response factor, putative [Ricinus communis] |