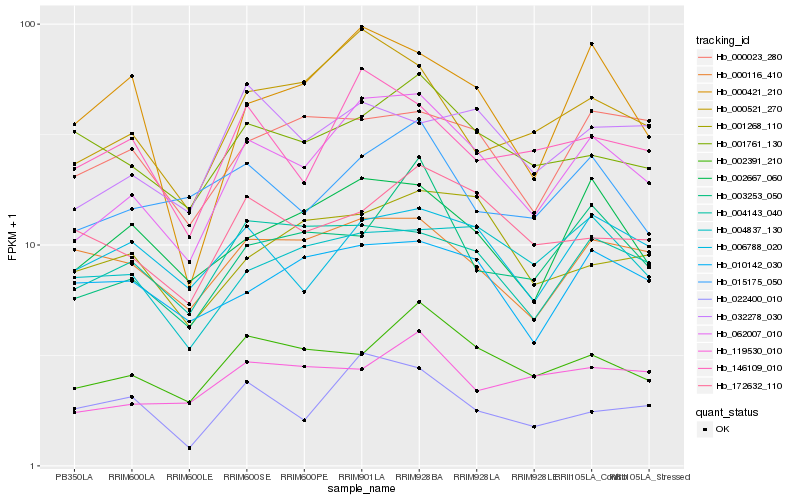
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_062007_010 |
0.0 |
- |
- |
PREDICTED: argininosuccinate lyase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000521_270 |
0.111563399 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 [Jatropha curcas] |
| 3 |
Hb_002667_060 |
0.117433181 |
- |
- |
mannose-1-phosphate guanyltransferase, putative [Ricinus communis] |
| 4 |
Hb_002391_210 |
0.1176209399 |
- |
- |
PREDICTED: putative F-box protein At1g65770 [Jatropha curcas] |
| 5 |
Hb_003253_050 |
0.1267608142 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 3 [Jatropha curcas] |
| 6 |
Hb_146109_010 |
0.1282199225 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004143_040 |
0.1293970702 |
- |
- |
argininosuccinate lyase, putative [Ricinus communis] |
| 8 |
Hb_032278_030 |
0.132384655 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 9 |
Hb_015175_050 |
0.1337039949 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 10 isoform X3 [Jatropha curcas] |
| 10 |
Hb_022400_010 |
0.1351692173 |
- |
- |
Nitrilase/cyanide hydratase and apolipoprotein N-acyltransferase family protein [Theobroma cacao] |
| 11 |
Hb_000421_210 |
0.1355396316 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_006788_020 |
0.1355649071 |
- |
- |
hypothetical protein JCGZ_21864 [Jatropha curcas] |
| 13 |
Hb_000023_280 |
0.1393183997 |
- |
- |
hypothetical protein POPTR_0006s28210g [Populus trichocarpa] |
| 14 |
Hb_000116_410 |
0.1401449265 |
- |
- |
hypothetical protein JCGZ_22416 [Jatropha curcas] |
| 15 |
Hb_119530_010 |
0.1402165554 |
- |
- |
- |
| 16 |
Hb_172632_110 |
0.1404733972 |
- |
- |
PREDICTED: pumilio homolog 5 [Jatropha curcas] |
| 17 |
Hb_001761_130 |
0.1421497476 |
- |
- |
PREDICTED: uncharacterized protein LOC105646118 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001268_110 |
0.1426186698 |
- |
- |
PREDICTED: uncharacterized protein LOC105630183 isoform X2 [Jatropha curcas] |
| 19 |
Hb_010142_030 |
0.143835223 |
- |
- |
PREDICTED: isoamylase 3, chloroplastic isoform X2 [Jatropha curcas] |
| 20 |
Hb_004837_130 |
0.1445097038 |
- |
- |
PREDICTED: armadillo repeat-containing protein 8 [Jatropha curcas] |