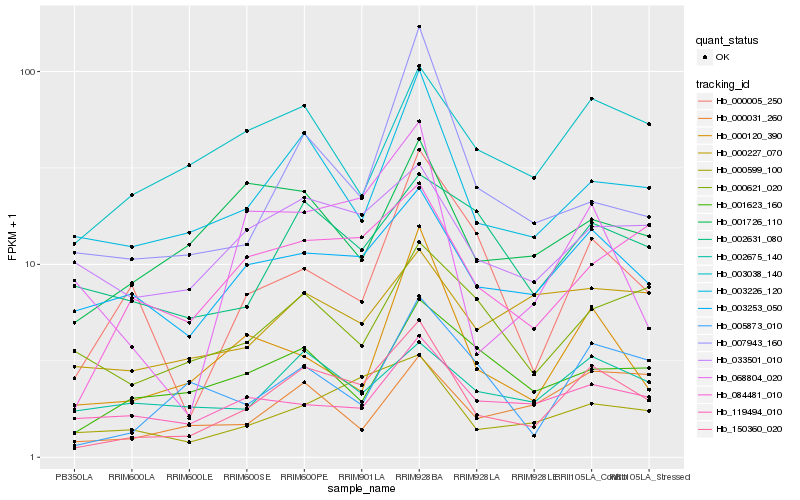
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_150360_020 |
0.0 |
- |
- |
PREDICTED: N-alpha-acetyltransferase 11 [Jatropha curcas] |
| 2 |
Hb_003226_120 |
0.1662929755 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000599_100 |
0.172253026 |
- |
- |
hypothetical protein B456_001G254000 [Gossypium raimondii] |
| 4 |
Hb_003253_050 |
0.1756752474 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 3 [Jatropha curcas] |
| 5 |
Hb_084481_010 |
0.1766773131 |
- |
- |
hypothetical protein POPTR_0011s036602g, partial [Populus trichocarpa] |
| 6 |
Hb_033501_010 |
0.1930153782 |
- |
- |
hypothetical protein JCGZ_21381 [Jatropha curcas] |
| 7 |
Hb_005873_010 |
0.194697615 |
- |
- |
cysteine desulfurylase, putative [Ricinus communis] |
| 8 |
Hb_003038_140 |
0.1999043077 |
- |
- |
soluble inorganic pyrophosphatase [Hevea brasiliensis] |
| 9 |
Hb_002675_140 |
0.2040982047 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 10 |
Hb_002631_080 |
0.2057933279 |
- |
- |
glyceraldehyde 3-phosphate dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_000031_260 |
0.2074671507 |
- |
- |
hypothetical protein POPTR_0018s03360g [Populus trichocarpa] |
| 12 |
Hb_068804_020 |
0.2083268204 |
- |
- |
hypothetical protein AMTR_s00046p00231970 [Amborella trichopoda] |
| 13 |
Hb_001726_110 |
0.2087032406 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 19 [Jatropha curcas] |
| 14 |
Hb_001623_160 |
0.2107616437 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At2g33170 [Jatropha curcas] |
| 15 |
Hb_000227_070 |
0.210888586 |
- |
- |
PREDICTED: ATP-dependent (S)-NAD(P)H-hydrate dehydratase isoform X2 [Jatropha curcas] |
| 16 |
Hb_000120_390 |
0.2114437371 |
- |
- |
PREDICTED: uncharacterized transporter YBR287W-like [Jatropha curcas] |
| 17 |
Hb_119494_010 |
0.2115078578 |
- |
- |
PREDICTED: box C/D snoRNA protein 1 [Jatropha curcas] |
| 18 |
Hb_000621_020 |
0.2115621197 |
- |
- |
PREDICTED: mannose-6-phosphate isomerase 1 [Jatropha curcas] |
| 19 |
Hb_007943_160 |
0.2115939051 |
- |
- |
PREDICTED: aspartate aminotransferase, cytoplasmic [Jatropha curcas] |
| 20 |
Hb_000005_250 |
0.2145256668 |
- |
- |
conserved hypothetical protein [Ricinus communis] |