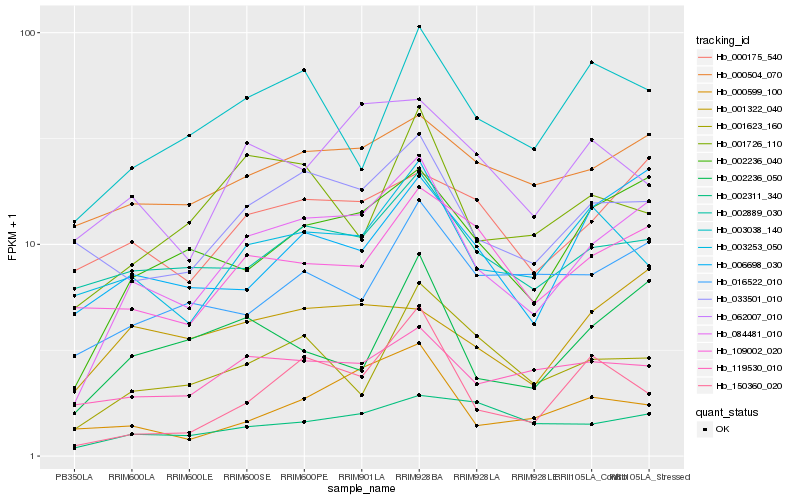
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_084481_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0011s036602g, partial [Populus trichocarpa] |
| 2 |
Hb_002236_040 |
0.1207696457 |
- |
- |
PREDICTED: cytochrome c [Jatropha curcas] |
| 3 |
Hb_033501_010 |
0.1542901478 |
- |
- |
hypothetical protein JCGZ_21381 [Jatropha curcas] |
| 4 |
Hb_000175_540 |
0.160433256 |
- |
- |
PREDICTED: uncharacterized protein LOC105634886 [Jatropha curcas] |
| 5 |
Hb_002889_030 |
0.1607018663 |
- |
- |
PREDICTED: uncharacterized protein At3g49140 [Jatropha curcas] |
| 6 |
Hb_000504_070 |
0.1622275199 |
- |
- |
PREDICTED: pescadillo homolog isoform X1 [Jatropha curcas] |
| 7 |
Hb_109002_020 |
0.1659268305 |
- |
- |
PREDICTED: CTP synthase-like [Jatropha curcas] |
| 8 |
Hb_006698_030 |
0.1667801452 |
- |
- |
PREDICTED: uncharacterized protein LOC105633969 [Jatropha curcas] |
| 9 |
Hb_002236_050 |
0.1692345864 |
- |
- |
- |
| 10 |
Hb_119530_010 |
0.1706374797 |
- |
- |
- |
| 11 |
Hb_003253_050 |
0.1731711737 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 3 [Jatropha curcas] |
| 12 |
Hb_000599_100 |
0.1734809988 |
- |
- |
hypothetical protein B456_001G254000 [Gossypium raimondii] |
| 13 |
Hb_002311_340 |
0.1735282836 |
- |
- |
phospholipase d zeta, putative [Ricinus communis] |
| 14 |
Hb_016522_010 |
0.174556876 |
- |
- |
PREDICTED: short-chain dehydrogenase/reductase 2b-like [Jatropha curcas] |
| 15 |
Hb_003038_140 |
0.1746685072 |
- |
- |
soluble inorganic pyrophosphatase [Hevea brasiliensis] |
| 16 |
Hb_001623_160 |
0.175804982 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At2g33170 [Jatropha curcas] |
| 17 |
Hb_001726_110 |
0.1763304664 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 19 [Jatropha curcas] |
| 18 |
Hb_150360_020 |
0.1766773131 |
- |
- |
PREDICTED: N-alpha-acetyltransferase 11 [Jatropha curcas] |
| 19 |
Hb_062007_010 |
0.1776506683 |
- |
- |
PREDICTED: argininosuccinate lyase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001322_040 |
0.1797473231 |
- |
- |
- |