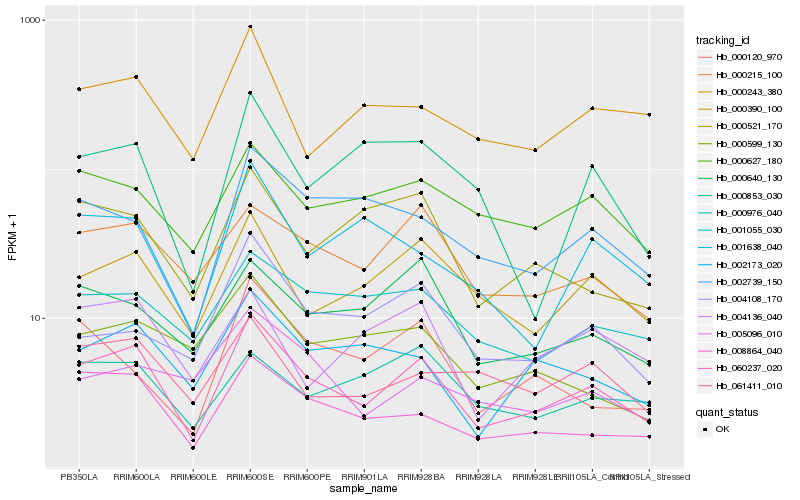
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_060237_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_008864_040 |
0.1431247491 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15980 [Jatropha curcas] |
| 3 |
Hb_000390_100 |
0.1502506042 |
- |
- |
alpha-hydroxynitrile lyase family protein [Populus trichocarpa] |
| 4 |
Hb_000215_100 |
0.1562359641 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002173_020 |
0.1615492714 |
- |
- |
PREDICTED: uncharacterized protein LOC105801078 [Gossypium raimondii] |
| 6 |
Hb_000627_180 |
0.1697093179 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001055_030 |
0.1698915826 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 14 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000640_130 |
0.1716635956 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_002739_150 |
0.1734647094 |
- |
- |
Uncharacterized protein TCM_016791 [Theobroma cacao] |
| 10 |
Hb_061411_010 |
0.1735110929 |
- |
- |
PREDICTED: ATPase WRNIP1 [Jatropha curcas] |
| 11 |
Hb_000243_380 |
0.1743522722 |
- |
- |
PREDICTED: histone H1 [Jatropha curcas] |
| 12 |
Hb_000521_170 |
0.1747756402 |
- |
- |
PREDICTED: callose synthase 1 [Jatropha curcas] |
| 13 |
Hb_001638_040 |
0.1754423559 |
- |
- |
Nucleobase-ascorbate transporter 3 [Glycine soja] |
| 14 |
Hb_004108_170 |
0.1817253293 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004136_040 |
0.1819501173 |
- |
- |
putative protein kinase [Arabidopsis thaliana] |
| 16 |
Hb_000853_030 |
0.1832605087 |
- |
- |
sucrose synthase 2 [Hevea brasiliensis] |
| 17 |
Hb_005096_010 |
0.1863376934 |
- |
- |
PREDICTED: uncharacterized protein LOC105641403 [Jatropha curcas] |
| 18 |
Hb_000976_040 |
0.1865719307 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 19 |
Hb_000599_130 |
0.1872304665 |
- |
- |
PREDICTED: U-box domain-containing protein 3 [Jatropha curcas] |
| 20 |
Hb_000120_970 |
0.1873431505 |
- |
- |
PREDICTED: probable acyl-activating enzyme 2 isoform X2 [Jatropha curcas] |