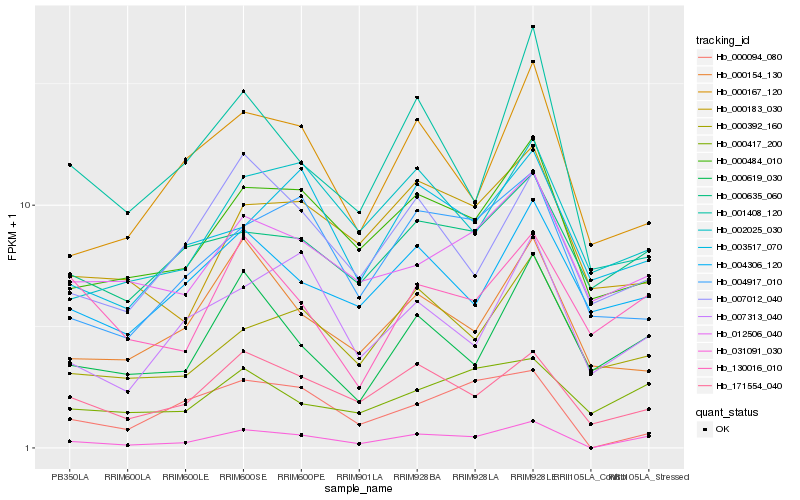
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_031091_030 |
0.0 |
- |
- |
hypothetical protein OsI_25921 [Oryza sativa Indica Group] |
| 2 |
Hb_171554_040 |
0.1820508774 |
- |
- |
PREDICTED: phytanoyl-CoA dioxygenase [Populus euphratica] |
| 3 |
Hb_000619_030 |
0.1908675956 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_000484_010 |
0.1914599916 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 23 [Jatropha curcas] |
| 5 |
Hb_000094_080 |
0.1929696695 |
- |
- |
PREDICTED: uncharacterized protein LOC105633792 [Jatropha curcas] |
| 6 |
Hb_007313_040 |
0.1986346476 |
- |
- |
hypothetical protein JCGZ_23401 [Jatropha curcas] |
| 7 |
Hb_000167_120 |
0.2008659668 |
- |
- |
PREDICTED: U-box domain-containing protein 14 [Jatropha curcas] |
| 8 |
Hb_000392_160 |
0.2025553747 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_007012_040 |
0.207393801 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 44 [Jatropha curcas] |
| 10 |
Hb_004917_010 |
0.2084181738 |
- |
- |
PREDICTED: U-box domain-containing protein 13-like [Jatropha curcas] |
| 11 |
Hb_002025_030 |
0.2094570827 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 12 |
Hb_012506_040 |
0.210262489 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105637346 [Jatropha curcas] |
| 13 |
Hb_000154_130 |
0.2103096014 |
- |
- |
PREDICTED: flowering time control protein FPA isoform X1 [Jatropha curcas] |
| 14 |
Hb_000417_200 |
0.2107743724 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: pentatricopeptide repeat-containing protein DOT4, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001408_120 |
0.2118315314 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 16 |
Hb_130016_010 |
0.2119157638 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000183_030 |
0.2123104413 |
- |
- |
PREDICTED: probable zinc transporter protein DDB_G0291141 [Populus euphratica] |
| 18 |
Hb_004306_120 |
0.2126178935 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_000635_060 |
0.2139070187 |
- |
- |
PREDICTED: uncharacterized protein LOC105644797 [Jatropha curcas] |
| 20 |
Hb_003517_070 |
0.2156815836 |
- |
- |
PREDICTED: protein transport protein Sec24-like At3g07100 [Jatropha curcas] |