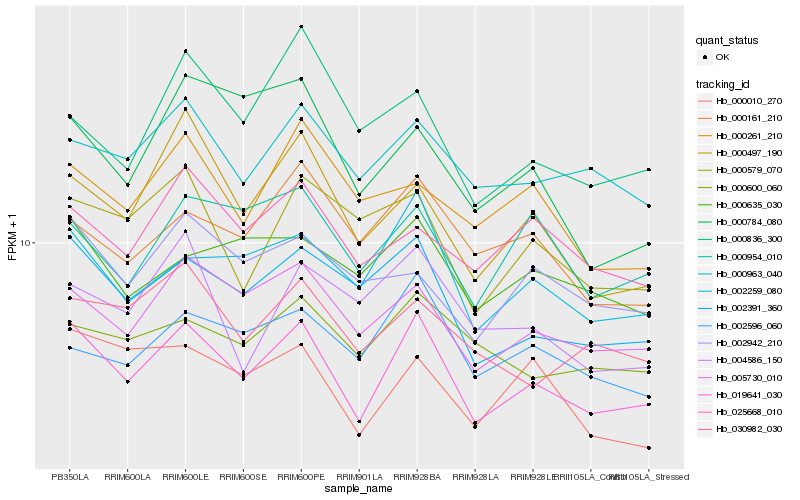
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_019641_030 |
0.0 |
- |
- |
chitin elicitor receptor kinase 1 [Chrysanthemum boreale] |
| 2 |
Hb_002391_360 |
0.1089625154 |
- |
- |
hypothetical protein CICLE_v10026208mg [Citrus clementina] |
| 3 |
Hb_005730_010 |
0.1110329603 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 7 isoform X1 [Populus euphratica] |
| 4 |
Hb_002259_080 |
0.1147609158 |
- |
- |
protein transport protein sec23, putative [Ricinus communis] |
| 5 |
Hb_000497_190 |
0.1258638047 |
- |
- |
sterol isomerase, putative [Ricinus communis] |
| 6 |
Hb_000963_040 |
0.1357832715 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 7 |
Hb_002596_060 |
0.1373142977 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000579_070 |
0.1377029046 |
- |
- |
hypothetical protein CISIN_1g0119542mg, partial [Citrus sinensis] |
| 9 |
Hb_000954_010 |
0.138453074 |
- |
- |
PREDICTED: uncharacterized protein LOC105646975 [Jatropha curcas] |
| 10 |
Hb_000161_210 |
0.1389656293 |
- |
- |
PREDICTED: uncharacterized protein At5g49945-like [Jatropha curcas] |
| 11 |
Hb_030982_030 |
0.1409194199 |
- |
- |
PREDICTED: uncharacterized protein LOC105636001 isoform X1 [Jatropha curcas] |
| 12 |
Hb_025668_010 |
0.1414700668 |
- |
- |
unnamed protein product [Coffea canephora] |
| 13 |
Hb_000010_270 |
0.1420383952 |
- |
- |
PREDICTED: vignain [Jatropha curcas] |
| 14 |
Hb_000600_060 |
0.1445723344 |
- |
- |
PREDICTED: uncharacterized protein LOC105633515 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000635_030 |
0.1446810026 |
- |
- |
mannosyl-oligosaccharide glucosidase, putative [Ricinus communis] |
| 16 |
Hb_002942_210 |
0.1448282993 |
- |
- |
alpha-2,8-sialyltransferase 8b, putative [Ricinus communis] |
| 17 |
Hb_000784_080 |
0.145445513 |
- |
- |
protein disulfide isomerase, putative [Ricinus communis] |
| 18 |
Hb_000836_300 |
0.1455368095 |
- |
- |
caax prenyl protease ste24, putative [Ricinus communis] |
| 19 |
Hb_004586_150 |
0.1456960565 |
- |
- |
PREDICTED: uncharacterized protein LOC105630688 [Jatropha curcas] |
| 20 |
Hb_000261_210 |
0.1472500035 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |