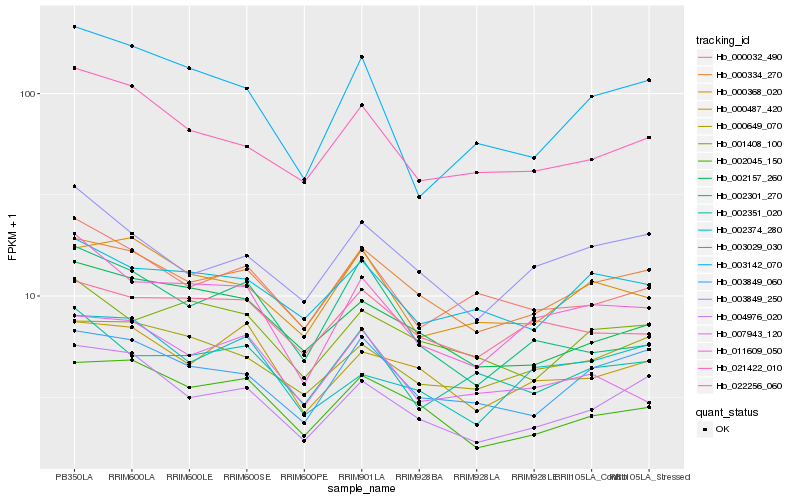
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011609_050 |
0.0 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_002351_020 |
0.0969304454 |
- |
- |
PREDICTED: rRNA methyltransferase 3B, mitochondrial [Jatropha curcas] |
| 3 |
Hb_003849_250 |
0.0983306741 |
- |
- |
PREDICTED: protein MAK16 homolog [Jatropha curcas] |
| 4 |
Hb_003142_070 |
0.0991807379 |
- |
- |
hypothetical protein F383_14657 [Gossypium arboreum] |
| 5 |
Hb_000334_270 |
0.0991976543 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_022256_060 |
0.1003579428 |
- |
- |
PREDICTED: uncharacterized protein LOC105628137 [Jatropha curcas] |
| 7 |
Hb_021422_010 |
0.1008827806 |
- |
- |
PREDICTED: alanine--glyoxylate aminotransferase 2 homolog 1, mitochondrial [Jatropha curcas] |
| 8 |
Hb_001408_100 |
0.1013635053 |
- |
- |
pseudouridylate synthase, putative [Ricinus communis] |
| 9 |
Hb_000032_490 |
0.101965615 |
- |
- |
PREDICTED: uncharacterized protein At4g10930-like [Jatropha curcas] |
| 10 |
Hb_004976_020 |
0.1032906081 |
- |
- |
PREDICTED: probable nucleolar protein 5-1 [Jatropha curcas] |
| 11 |
Hb_002157_260 |
0.1034747086 |
- |
- |
PREDICTED: partner of Y14 and mago-like [Jatropha curcas] |
| 12 |
Hb_003029_030 |
0.1069325388 |
- |
- |
PREDICTED: uncharacterized protein LOC105642632 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000649_070 |
0.1079343489 |
- |
- |
PREDICTED: probable UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase 2 [Jatropha curcas] |
| 14 |
Hb_007943_120 |
0.1091883076 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 5 isoform X4 [Jatropha curcas] |
| 15 |
Hb_000487_420 |
0.1096674216 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At1g04390 [Jatropha curcas] |
| 16 |
Hb_000368_020 |
0.1111971904 |
- |
- |
uroporphyrin-III methyltransferase, putative [Ricinus communis] |
| 17 |
Hb_003849_060 |
0.1117636159 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g13040, mitochondrial [Jatropha curcas] |
| 18 |
Hb_002374_280 |
0.1119844531 |
- |
- |
- |
| 19 |
Hb_002301_270 |
0.1121249963 |
- |
- |
PREDICTED: glyoxysomal processing protease, glyoxysomal isoform X1 [Jatropha curcas] |
| 20 |
Hb_002045_150 |
0.1130623168 |
- |
- |
histone acetyltransferase gcn5, putative [Ricinus communis] |