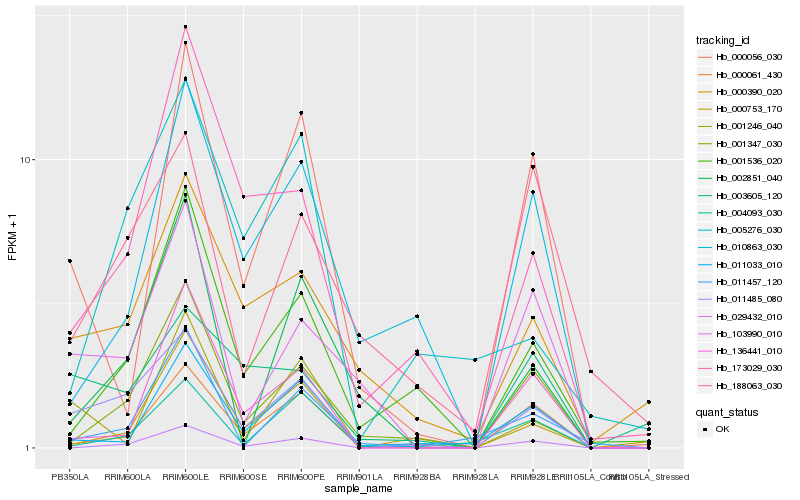
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011485_080 |
0.0 |
- |
- |
PREDICTED: 50S ribosomal protein L29, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001347_030 |
0.224900306 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 3 |
Hb_000390_020 |
0.2311093425 |
transcription factor |
TF Family: GRF |
PREDICTED: growth-regulating factor 4-like [Jatropha curcas] |
| 4 |
Hb_173029_030 |
0.2332258908 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_103990_010 |
0.2369853074 |
- |
- |
PREDICTED: light-induced protein, chloroplastic-like [Nicotiana sylvestris] |
| 6 |
Hb_000061_430 |
0.2375798084 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001536_020 |
0.2436930838 |
transcription factor |
TF Family: GRF |
PREDICTED: uncharacterized protein LOC105631006 [Jatropha curcas] |
| 8 |
Hb_029432_010 |
0.2442189694 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 9 |
Hb_003605_120 |
0.2559493604 |
- |
- |
hypothetical protein POPTR_0013s12060g [Populus trichocarpa] |
| 10 |
Hb_005276_030 |
0.2582631314 |
- |
- |
PREDICTED: probable inactive patatin-like protein 9 [Jatropha curcas] |
| 11 |
Hb_000753_170 |
0.2588162621 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_011033_010 |
0.2647668023 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 13 |
Hb_136441_010 |
0.2680674919 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 14 |
Hb_001246_040 |
0.270197657 |
- |
- |
hypothetical protein JCGZ_14673 [Jatropha curcas] |
| 15 |
Hb_000056_030 |
0.2723454015 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g14390 isoform X1 [Jatropha curcas] |
| 16 |
Hb_004093_030 |
0.275305014 |
- |
- |
hypothetical protein RCOM_1406750 [Ricinus communis] |
| 17 |
Hb_188063_030 |
0.2766442484 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 18 |
Hb_002851_040 |
0.2770389755 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 5 [Jatropha curcas] |
| 19 |
Hb_011457_120 |
0.2777084966 |
- |
- |
PREDICTED: sulfate transporter 3.1-like [Jatropha curcas] |
| 20 |
Hb_010863_030 |
0.2801980868 |
- |
- |
PREDICTED: uncharacterized protein LOC105642466 [Jatropha curcas] |