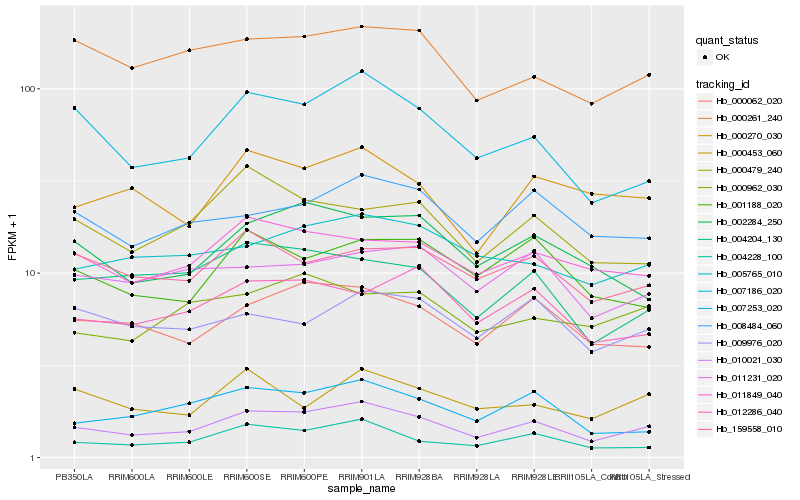
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010021_030 |
0.0 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40A isoform X4 [Jatropha curcas] |
| 2 |
Hb_007186_020 |
0.088786397 |
- |
- |
Eukaryotic initiation factor 4A-3 [Gossypium arboreum] |
| 3 |
Hb_000062_020 |
0.0966167483 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 4 |
Hb_004228_100 |
0.0999384235 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 5 |
Hb_000270_030 |
0.1072533968 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0015s07640g [Populus trichocarpa] |
| 6 |
Hb_000261_240 |
0.1072874119 |
- |
- |
RecName: Full=Elongation factor 1-alpha; Short=EF-1-alpha [Manihot esculenta] |
| 7 |
Hb_008484_060 |
0.1078050246 |
- |
- |
PREDICTED: allantoate deiminase isoform X3 [Populus euphratica] |
| 8 |
Hb_004204_130 |
0.1086256699 |
- |
- |
PREDICTED: uncharacterized protein LOC103930912 [Pyrus x bretschneideri] |
| 9 |
Hb_007253_020 |
0.1092019147 |
- |
- |
PREDICTED: putative deoxyribonuclease TATDN1 [Jatropha curcas] |
| 10 |
Hb_011849_040 |
0.1101220244 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 11 |
Hb_005765_010 |
0.1111652321 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002284_250 |
0.1144041625 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 13 |
Hb_159558_010 |
0.1148283964 |
- |
- |
PREDICTED: uncharacterized protein LOC105635846 [Jatropha curcas] |
| 14 |
Hb_012286_040 |
0.1151150454 |
- |
- |
PREDICTED: pre-mRNA-splicing factor RSE1 [Jatropha curcas] |
| 15 |
Hb_001188_020 |
0.1163966333 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000453_060 |
0.1167778232 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g11690 [Jatropha curcas] |
| 17 |
Hb_000479_240 |
0.1180751644 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |
| 18 |
Hb_011231_020 |
0.1182614012 |
- |
- |
PREDICTED: proline-, glutamic acid- and leucine-rich protein 1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_009976_020 |
0.1183451925 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105631286 [Jatropha curcas] |
| 20 |
Hb_000962_030 |
0.1186205048 |
- |
- |
PREDICTED: metal tolerance protein C2 [Jatropha curcas] |