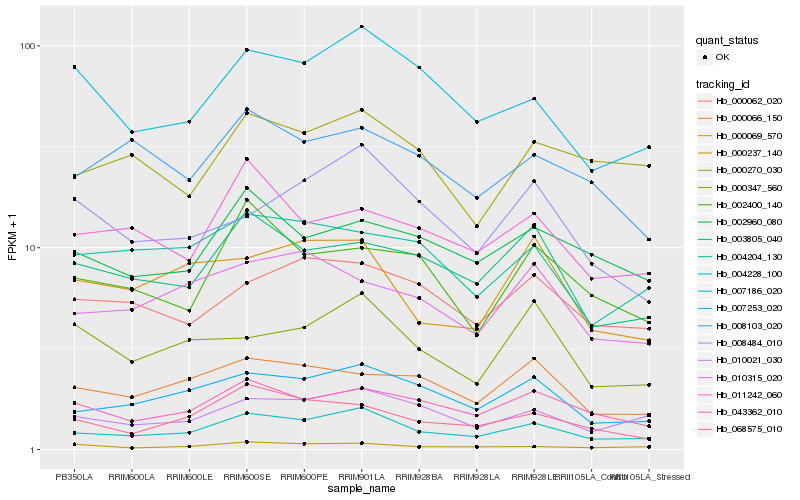
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004228_100 |
0.0 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 2 |
Hb_007253_020 |
0.0892536854 |
- |
- |
PREDICTED: putative deoxyribonuclease TATDN1 [Jatropha curcas] |
| 3 |
Hb_010021_030 |
0.0999384235 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40A isoform X4 [Jatropha curcas] |
| 4 |
Hb_007186_020 |
0.1089424528 |
- |
- |
Eukaryotic initiation factor 4A-3 [Gossypium arboreum] |
| 5 |
Hb_043362_010 |
0.1096008415 |
- |
- |
hypothetical protein JCGZ_23765 [Jatropha curcas] |
| 6 |
Hb_068575_010 |
0.1156607294 |
- |
- |
PREDICTED: protein GAMETE EXPRESSED 2 [Jatropha curcas] |
| 7 |
Hb_003805_040 |
0.1199173766 |
- |
- |
Sacsin [Glycine soja] |
| 8 |
Hb_000062_020 |
0.1262640109 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 9 |
Hb_008484_010 |
0.1275148383 |
- |
- |
hypothetical protein B456_008G050700 [Gossypium raimondii] |
| 10 |
Hb_000237_140 |
0.1276270892 |
- |
- |
PREDICTED: RNA-binding protein 28 [Jatropha curcas] |
| 11 |
Hb_000069_570 |
0.1281800168 |
- |
- |
PREDICTED: uncharacterized protein At5g05190-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000066_150 |
0.1286861725 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 13 |
Hb_002960_080 |
0.1295614908 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105635322 [Jatropha curcas] |
| 14 |
Hb_010315_020 |
0.1302276184 |
- |
- |
PREDICTED: uncharacterized protein LOC105640834 [Jatropha curcas] |
| 15 |
Hb_000270_030 |
0.1316371261 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0015s07640g [Populus trichocarpa] |
| 16 |
Hb_002400_140 |
0.1316550602 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 17 |
Hb_011242_060 |
0.1325727467 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 18 |
Hb_000347_560 |
0.1329902127 |
- |
- |
- |
| 19 |
Hb_008103_020 |
0.133654691 |
- |
- |
hypothetical protein POPTR_0003s20310g [Populus trichocarpa] |
| 20 |
Hb_004204_130 |
0.1336784555 |
- |
- |
PREDICTED: uncharacterized protein LOC103930912 [Pyrus x bretschneideri] |