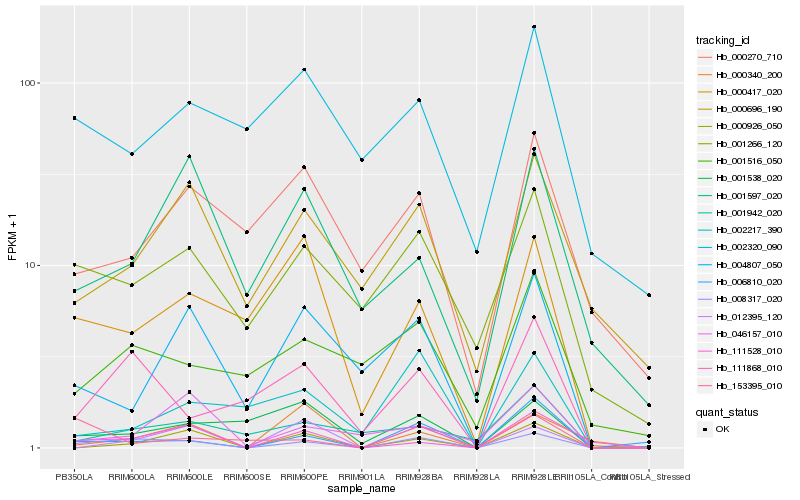
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008317_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_111868_010 |
0.2771646031 |
- |
- |
hypothetical protein CICLE_v10029531mg [Citrus clementina] |
| 3 |
Hb_001266_120 |
0.2908062736 |
- |
- |
auxilin, putative [Ricinus communis] |
| 4 |
Hb_006810_020 |
0.2912231246 |
- |
- |
PREDICTED: NADPH:quinone oxidoreductase-like [Jatropha curcas] |
| 5 |
Hb_111528_010 |
0.2967111702 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 2-like [Jatropha curcas] |
| 6 |
Hb_000926_050 |
0.3098762669 |
- |
- |
PREDICTED: vinorine synthase-like [Populus euphratica] |
| 7 |
Hb_000417_020 |
0.3111927835 |
- |
- |
PREDICTED: uncharacterized protein LOC105649674 [Jatropha curcas] |
| 8 |
Hb_012395_120 |
0.314035234 |
- |
- |
- |
| 9 |
Hb_000696_190 |
0.3197266678 |
- |
- |
PREDICTED: protein MID1-COMPLEMENTING ACTIVITY 1 [Jatropha curcas] |
| 10 |
Hb_001516_050 |
0.3215004686 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD3-1 [Jatropha curcas] |
| 11 |
Hb_004807_050 |
0.3229615459 |
- |
- |
hypothetical protein POPTR_0004s17300g [Populus trichocarpa] |
| 12 |
Hb_001538_020 |
0.3376399167 |
- |
- |
PREDICTED: uncharacterized protein LOC105629608 [Jatropha curcas] |
| 13 |
Hb_000340_200 |
0.3405985975 |
- |
- |
PREDICTED: uncharacterized protein LOC105650039 [Jatropha curcas] |
| 14 |
Hb_001942_020 |
0.341156212 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP16-3, chloroplastic [Jatropha curcas] |
| 15 |
Hb_002217_390 |
0.3424014016 |
- |
- |
PREDICTED: galactokinase [Jatropha curcas] |
| 16 |
Hb_153395_010 |
0.3427874158 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_001597_020 |
0.3434713338 |
- |
- |
PREDICTED: uncharacterized protein LOC105629846 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000270_710 |
0.3440750244 |
- |
- |
PREDICTED: WAT1-related protein At5g40240-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_046157_010 |
0.3449583869 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 2-like [Jatropha curcas] |
| 20 |
Hb_002320_090 |
0.3462402643 |
- |
- |
PREDICTED: endoglucanase 25-like [Jatropha curcas] |