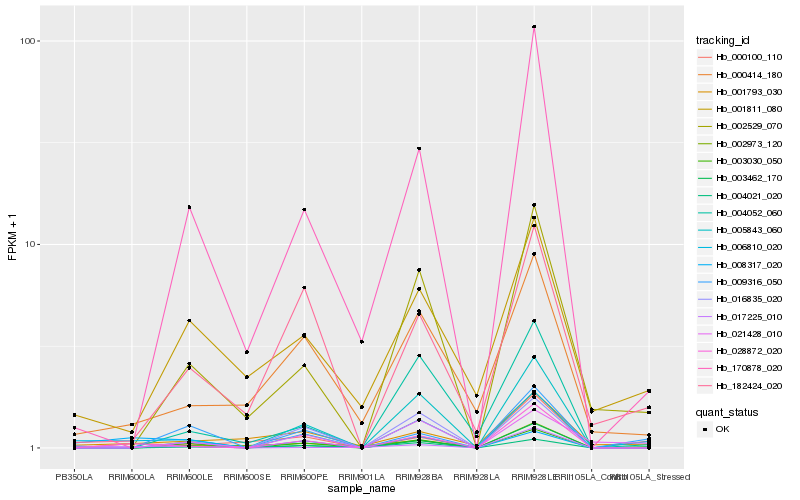
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006810_020 |
0.0 |
- |
- |
PREDICTED: NADPH:quinone oxidoreductase-like [Jatropha curcas] |
| 2 |
Hb_028872_020 |
0.254240182 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 3 |
Hb_004021_020 |
0.2716548709 |
- |
- |
PREDICTED: transposon Tf2-1 polyprotein isoform X1 [Zea mays] |
| 4 |
Hb_002529_070 |
0.2741466097 |
- |
- |
PREDICTED: putative late blight resistance protein homolog R1B-19 [Pyrus x bretschneideri] |
| 5 |
Hb_001793_030 |
0.2748387217 |
- |
- |
Cysteine-rich RLK (RECEPTOR-like protein kinase) 8 [Theobroma cacao] |
| 6 |
Hb_000100_110 |
0.2763653854 |
- |
- |
- |
| 7 |
Hb_000414_180 |
0.2783236558 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 isoform X2 [Vitis vinifera] |
| 8 |
Hb_182424_020 |
0.2788596011 |
- |
- |
hypothetical protein POPTR_0006s24190g [Populus trichocarpa] |
| 9 |
Hb_002973_120 |
0.2814582757 |
- |
- |
Uncharacterized protein TCM_024268 [Theobroma cacao] |
| 10 |
Hb_003030_050 |
0.2822271875 |
- |
- |
- |
| 11 |
Hb_005843_060 |
0.282462768 |
- |
- |
PREDICTED: protein FAR-RED IMPAIRED RESPONSE 1 isoform X3 [Nelumbo nucifera] |
| 12 |
Hb_170878_020 |
0.2825103147 |
- |
- |
- |
| 13 |
Hb_021428_010 |
0.2880843758 |
- |
- |
PREDICTED: putative cysteine-rich receptor-like protein kinase 34 [Jatropha curcas] |
| 14 |
Hb_016835_020 |
0.2883458582 |
- |
- |
unnamed protein product [Coffea canephora] |
| 15 |
Hb_004052_060 |
0.2886090025 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 16 |
Hb_017225_010 |
0.2888115764 |
- |
- |
PREDICTED: nodulation receptor kinase-like [Jatropha curcas] |
| 17 |
Hb_001811_080 |
0.289548465 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 18 |
Hb_009316_050 |
0.2906602893 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 19 |
Hb_003462_170 |
0.290807048 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_008317_020 |
0.2912231246 |
- |
- |
- |