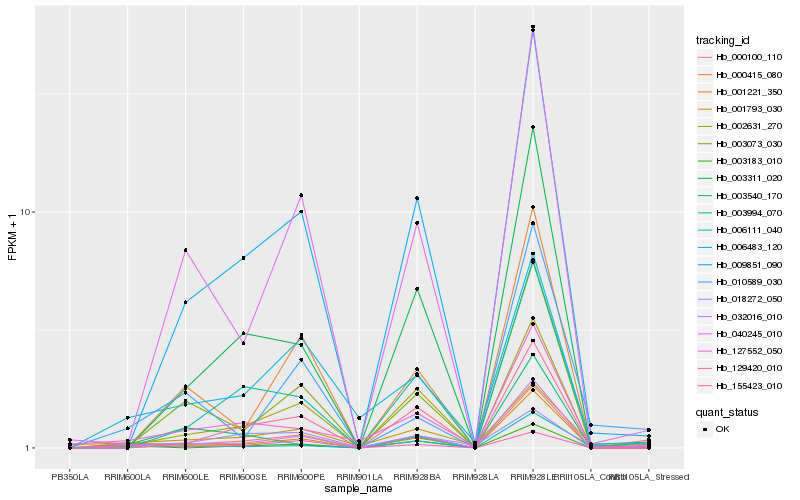
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000100_110 |
0.0 |
- |
- |
- |
| 2 |
Hb_001221_350 |
0.1842115215 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_006111_040 |
0.2071426197 |
- |
- |
PREDICTED: uncharacterized protein LOC103485923 [Cucumis melo] |
| 4 |
Hb_001793_030 |
0.2190655769 |
- |
- |
Cysteine-rich RLK (RECEPTOR-like protein kinase) 8 [Theobroma cacao] |
| 5 |
Hb_009851_090 |
0.219101109 |
- |
- |
Protein ABIL2, putative [Ricinus communis] |
| 6 |
Hb_127552_050 |
0.2247616606 |
- |
- |
hypothetical protein VITISV_030752 [Vitis vinifera] |
| 7 |
Hb_002631_270 |
0.2251210156 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0005s09930g [Populus trichocarpa] |
| 8 |
Hb_003183_010 |
0.2276758638 |
- |
- |
PREDICTED: uncharacterized protein LOC105785048, partial [Gossypium raimondii] |
| 9 |
Hb_032016_010 |
0.2298911328 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At3g28040-like isoform X1 [Citrus sinensis] |
| 10 |
Hb_006483_120 |
0.231806803 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_040245_010 |
0.2370162217 |
- |
- |
hypothetical protein CISIN_1g039575mg, partial [Citrus sinensis] |
| 12 |
Hb_010589_030 |
0.2373618186 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK2 [Jatropha curcas] |
| 13 |
Hb_018272_050 |
0.2378613787 |
- |
- |
hypothetical protein VITISV_037409 [Vitis vinifera] |
| 14 |
Hb_003311_020 |
0.2391720555 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_129420_010 |
0.2415502926 |
transcription factor |
TF Family: NAC |
NAC transcription factor 059 [Jatropha curcas] |
| 16 |
Hb_155423_010 |
0.2418687831 |
- |
- |
- |
| 17 |
Hb_000415_080 |
0.241877207 |
- |
- |
- |
| 18 |
Hb_003073_030 |
0.2425005451 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003540_170 |
0.2431876022 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21705, mitochondrial-like [Jatropha curcas] |
| 20 |
Hb_003994_070 |
0.2436512903 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |