| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
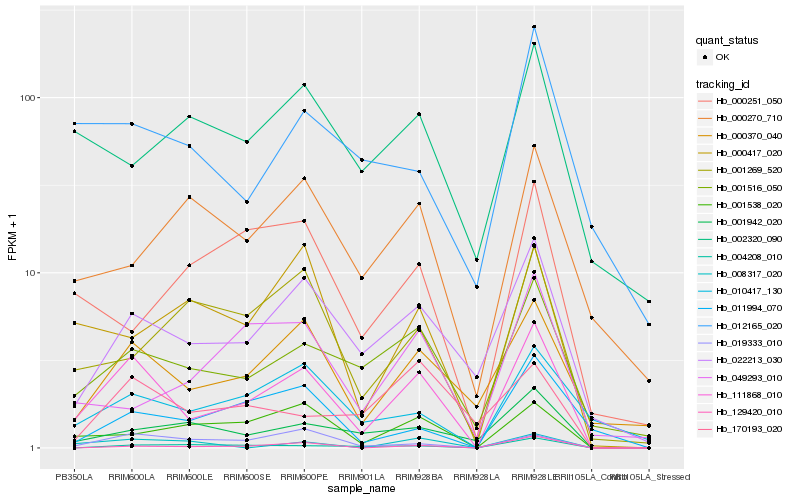
Hb_111868_010 |
0.0 |
- |
- |
hypothetical protein CICLE_v10029531mg [Citrus clementina] |
| 2 |
Hb_000370_040 |
0.2137191542 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 3 |
Hb_001516_050 |
0.215779378 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD3-1 [Jatropha curcas] |
| 4 |
Hb_022213_030 |
0.2176126575 |
- |
- |
PREDICTED: uncharacterized protein LOC105114631 [Populus euphratica] |
| 5 |
Hb_001538_020 |
0.2472360045 |
- |
- |
PREDICTED: uncharacterized protein LOC105629608 [Jatropha curcas] |
| 6 |
Hb_019333_010 |
0.2533250581 |
- |
- |
PREDICTED: uncharacterized protein LOC105633210 [Jatropha curcas] |
| 7 |
Hb_010417_130 |
0.2544959831 |
- |
- |
PREDICTED: probable membrane-associated kinase regulator 2 [Jatropha curcas] |
| 8 |
Hb_000417_020 |
0.2568336017 |
- |
- |
PREDICTED: uncharacterized protein LOC105649674 [Jatropha curcas] |
| 9 |
Hb_129420_010 |
0.2595452188 |
transcription factor |
TF Family: NAC |
NAC transcription factor 059 [Jatropha curcas] |
| 10 |
Hb_001942_020 |
0.2604790859 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP16-3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001269_520 |
0.2655078049 |
- |
- |
PREDICTED: putative receptor-like protein kinase At1g80870 [Jatropha curcas] |
| 12 |
Hb_011994_070 |
0.2669201867 |
- |
- |
PREDICTED: ras-related protein RABC2a-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_004208_010 |
0.2703365407 |
- |
- |
PREDICTED: uncharacterized protein LOC105914588 [Setaria italica] |
| 14 |
Hb_170193_020 |
0.2748178256 |
- |
- |
- |
| 15 |
Hb_008317_020 |
0.2771646031 |
- |
- |
- |
| 16 |
Hb_049293_010 |
0.2802207543 |
- |
- |
kinase, putative [Ricinus communis] |
| 17 |
Hb_012165_020 |
0.2804695973 |
- |
- |
hypothetical protein POPTR_0007s05770g [Populus trichocarpa] |
| 18 |
Hb_002320_090 |
0.2836476951 |
- |
- |
PREDICTED: endoglucanase 25-like [Jatropha curcas] |
| 19 |
Hb_000270_710 |
0.2850211063 |
- |
- |
PREDICTED: WAT1-related protein At5g40240-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_000251_050 |
0.2865734778 |
- |
- |
hypothetical protein JCGZ_21678 [Jatropha curcas] |