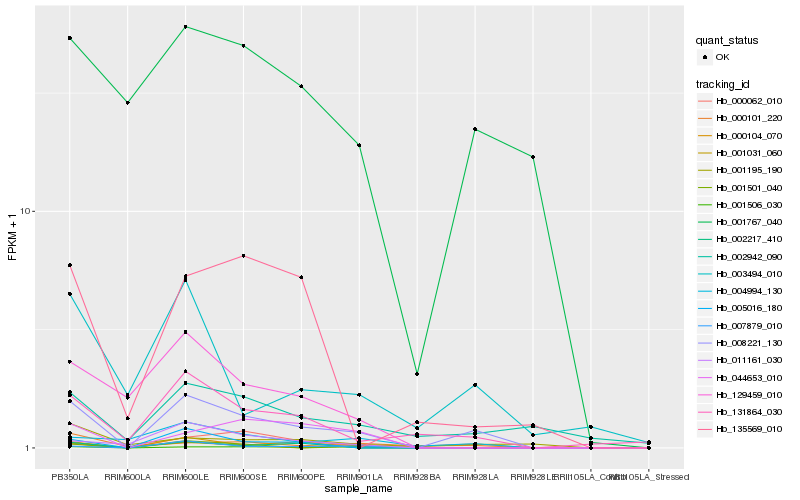
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008221_130 |
0.0 |
- |
- |
hypothetical protein VITISV_016971 [Vitis vinifera] |
| 2 |
Hb_131864_030 |
0.2697533355 |
transcription factor |
TF Family: C2H2 |
transcription factor, putative [Ricinus communis] |
| 3 |
Hb_000062_010 |
0.2946426826 |
- |
- |
Alpha-L-fucosidase 2 precursor, putative [Ricinus communis] |
| 4 |
Hb_004994_130 |
0.2958150851 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-like protein M [Jatropha curcas] |
| 5 |
Hb_007879_010 |
0.2997816374 |
- |
- |
hypothetical protein JCGZ_11284 [Jatropha curcas] |
| 6 |
Hb_002942_090 |
0.3025505754 |
- |
- |
DNAJ heat shock N-terminal domain-containing protein, putative [Theobroma cacao] |
| 7 |
Hb_000101_220 |
0.3029771899 |
- |
- |
anthranilate synthase component I, putative [Ricinus communis] |
| 8 |
Hb_001767_040 |
0.3031014974 |
- |
- |
dehydroascorbate reductase, putative [Ricinus communis] |
| 9 |
Hb_001506_030 |
0.3057857053 |
- |
- |
DNA (cytosine-5)-methyltransferase, putative [Ricinus communis] |
| 10 |
Hb_001195_190 |
0.3132906666 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003494_010 |
0.3133302949 |
- |
- |
hypothetical protein JCGZ_19838 [Jatropha curcas] |
| 12 |
Hb_129459_010 |
0.3171363478 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18840-like [Populus euphratica] |
| 13 |
Hb_005016_180 |
0.3188217634 |
- |
- |
phosphoglycerate kinase, putative [Ricinus communis] |
| 14 |
Hb_044653_010 |
0.3224402038 |
- |
- |
PREDICTED: GTPase Era, mitochondrial [Jatropha curcas] |
| 15 |
Hb_002217_410 |
0.3246904667 |
- |
- |
hypothetical protein L484_027883 [Morus notabilis] |
| 16 |
Hb_011161_030 |
0.3266514584 |
- |
- |
PREDICTED: uncharacterized protein LOC105638906 [Jatropha curcas] |
| 17 |
Hb_001501_040 |
0.3276172228 |
- |
- |
PREDICTED: uncharacterized protein LOC102663792 [Glycine max] |
| 18 |
Hb_000104_070 |
0.3322329677 |
- |
- |
PREDICTED: citrate-binding protein-like [Jatropha curcas] |
| 19 |
Hb_135569_010 |
0.3340194668 |
- |
- |
PREDICTED: uncharacterized protein LOC105108579 [Populus euphratica] |
| 20 |
Hb_001031_060 |
0.3345857697 |
- |
- |
PREDICTED: uncharacterized protein LOC103940329 [Pyrus x bretschneideri] |